You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001790_00202
You are here: Home > Sequence: MGYG000001790_00202
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
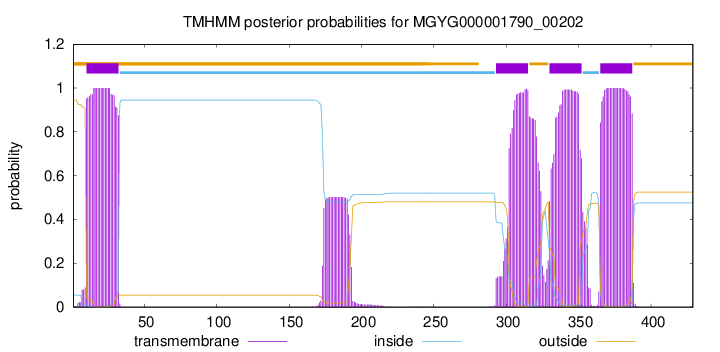
TMHMM annotations
Basic Information help
| Species | Phascolarctobacterium_A sp900553055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Acidaminococcales; Acidaminococcaceae; Phascolarctobacterium_A; Phascolarctobacterium_A sp900553055 | |||||||||||
| CAZyme ID | MGYG000001790_00202 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4958; End: 6247 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 47 | 269 | 3.2e-32 | 0.9739130434782609 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06438 | EpsO_like | 3.53e-93 | 52 | 231 | 4 | 183 | EpsO protein participates in the methanolan synthesis. The Methylobacillus sp EpsO protein is predicted to participate in the methanolan synthesis. Methanolan is an exopolysaccharide (EPS), composed of glucose, mannose and galactose. A 21 genes cluster was predicted to participate in the methanolan synthesis. Gene disruption analysis revealed that EpsO is one of the glycosyltransferase enzymes involved in the synthesis of repeating sugar units onto the lipid carrier. |
| COG1215 | BcsA | 6.79e-42 | 7 | 381 | 16 | 394 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| cd06423 | CESA_like | 9.39e-40 | 53 | 228 | 5 | 180 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| pfam13641 | Glyco_tranf_2_3 | 1.19e-26 | 46 | 271 | 3 | 230 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
| cd06436 | GlcNAc-1-P_transferase | 9.73e-14 | 49 | 228 | 2 | 191 | N-acetyl-glucosamine transferase is involved in the synthesis of Poly-beta-1,6-N-acetyl-D-glucosamine. N-acetyl-glucosamine transferase is responsible for the synthesis of bacteria Poly-beta-1,6-N-acetyl-D-glucosamine (PGA). Poly-beta-1,6-N-acetyl-D-glucosamine is a homopolymer that serves as an adhesion for the maintenance of biofilm structural stability in diverse eubacteria. N-acetyl-glucosamine transferase is the product of gene pgaC. Genetic analysis indicated that all four genes of the pgaABCD locus were required for the PGA production, pgaC being a glycosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTV77611.1 | 6.28e-308 | 1 | 426 | 1 | 426 |
| BBG64019.1 | 1.39e-246 | 1 | 426 | 1 | 425 |
| QNP76606.1 | 1.97e-246 | 1 | 426 | 1 | 425 |
| ADB47159.1 | 1.37e-216 | 1 | 429 | 1 | 428 |
| AEQ22382.1 | 2.88e-212 | 5 | 428 | 5 | 427 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q3MB01 | 2.43e-12 | 48 | 286 | 109 | 347 | Beta-monoglucosyldiacylglycerol synthase OS=Trichormus variabilis (strain ATCC 29413 / PCC 7937) OX=240292 GN=Ava_2217 PE=1 SV=1 |
| Q8YMK0 | 2.43e-12 | 48 | 286 | 109 | 347 | Beta-monoglucosyldiacylglycerol synthase OS=Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576) OX=103690 GN=all4933 PE=1 SV=1 |
| D4GU74 | 1.41e-07 | 42 | 258 | 41 | 240 | Low-salt glycan biosynthesis hexosyltransferase Agl6 OS=Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) OX=309800 GN=agl6 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
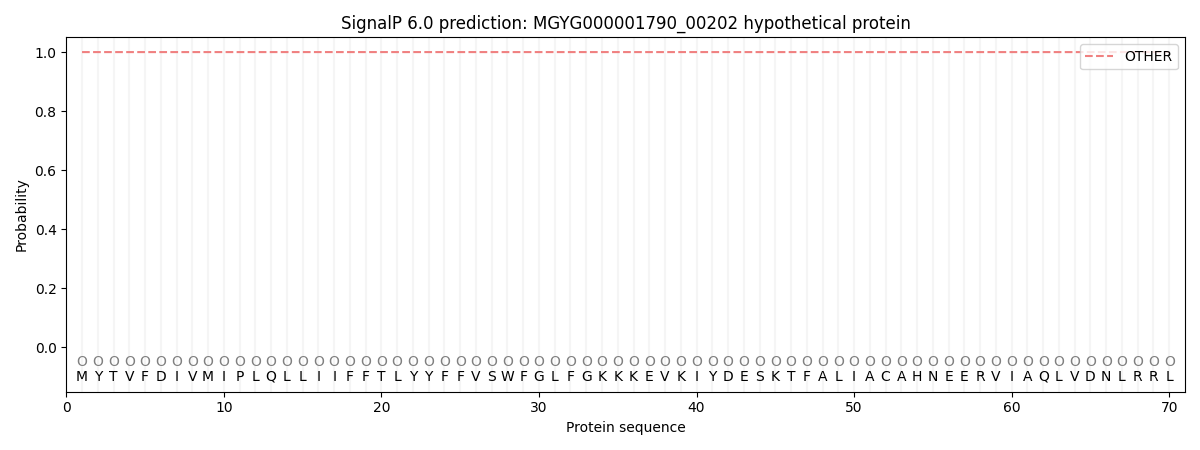
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000029 | 0.000005 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |