You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001804_00193
You are here: Home > Sequence: MGYG000001804_00193
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; CAG-831; | |||||||||||
| CAZyme ID | MGYG000001804_00193 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9995; End: 11239 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 85 | 391 | 3.3e-80 | 0.9239766081871345 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 3.32e-94 | 75 | 390 | 1 | 326 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam13088 | BNR_2 | 1.22e-22 | 122 | 392 | 15 | 278 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam13859 | BNR_3 | 8.01e-12 | 102 | 304 | 8 | 212 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| COG4409 | NanH | 8.11e-12 | 91 | 386 | 274 | 639 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam15902 | Sortilin-Vps10 | 3.31e-05 | 249 | 359 | 6 | 110 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFZ55957.1 | 3.45e-137 | 76 | 404 | 28 | 359 |
| ASO05493.1 | 1.53e-131 | 72 | 391 | 29 | 348 |
| AUC86605.1 | 6.92e-131 | 73 | 395 | 23 | 348 |
| QXD15092.1 | 2.59e-129 | 72 | 391 | 30 | 349 |
| APQ16737.1 | 4.20e-129 | 72 | 391 | 31 | 350 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6MNJ_A | 3.47e-42 | 92 | 387 | 200 | 521 | Hadzamicrobial sialidase Hz136 [Alistipes],6MNJ_B Hadza microbial sialidase Hz136 [Alistipes] |
| 1EUR_A | 3.81e-40 | 92 | 387 | 25 | 344 | Sialidase[Micromonospora viridifaciens],1EUS_A Sialidase Complexed With 2-Deoxy-2,3-Dehydro-N- Acetylneuraminic Acid [Micromonospora viridifaciens] |
| 7LBU_A | 5.48e-39 | 92 | 390 | 94 | 416 | ChainA, Exo-alpha-sialidase [Cutibacterium acnes],7LBV_A Chain A, Exo-alpha-sialidase [Cutibacterium acnes] |
| 1W8N_A | 6.99e-39 | 92 | 387 | 21 | 340 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
| 1EUT_A | 1.36e-38 | 92 | 387 | 25 | 344 | Sialidase,Large 68kd Form, Complexed With Galactose [Micromonospora viridifaciens],1EUU_A Sialidase Or Neuraminidase, Large 68kd Form [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 1.05e-37 | 92 | 387 | 67 | 386 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P31206 | 8.06e-32 | 90 | 387 | 201 | 526 | Sialidase OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=nanH PE=3 SV=2 |
| A5PF10 | 2.50e-28 | 88 | 406 | 74 | 414 | Sialidase-1 OS=Sus scrofa OX=9823 GN=NEU1 PE=3 SV=1 |
| A6BMK7 | 3.39e-28 | 88 | 388 | 73 | 395 | Sialidase-1 OS=Bos taurus OX=9913 GN=NEU1 PE=2 SV=2 |
| Q99519 | 2.25e-27 | 88 | 406 | 73 | 413 | Sialidase-1 OS=Homo sapiens OX=9606 GN=NEU1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
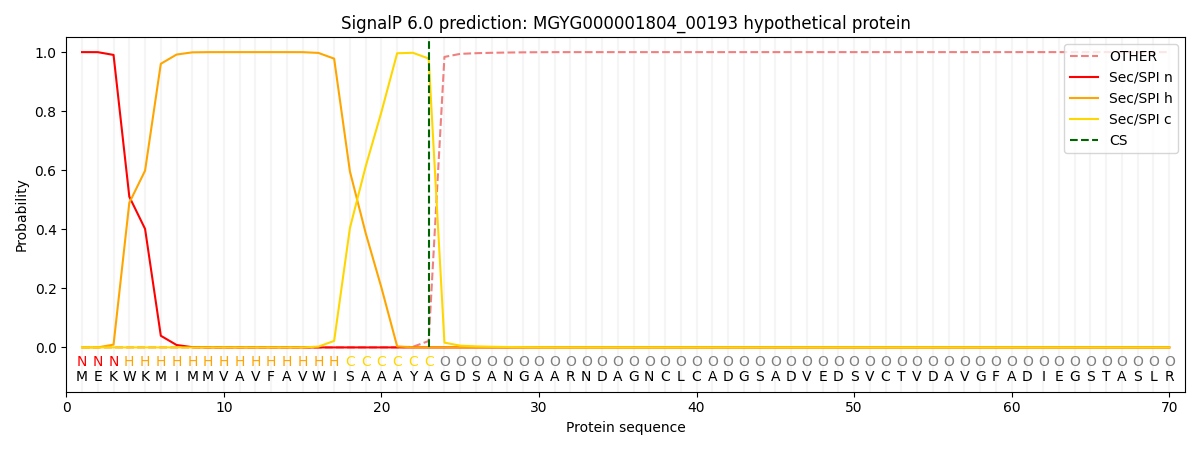
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000330 | 0.998929 | 0.000211 | 0.000173 | 0.000163 | 0.000160 |
