You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001805_00556
You are here: Home > Sequence: MGYG000001805_00556
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000001805_00556 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13165; End: 14487 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 49 | 315 | 1.7e-41 | 0.8368055555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 7.18e-13 | 51 | 289 | 100 | 348 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02432 | PLN02432 | 1.39e-11 | 169 | 359 | 111 | 290 | putative pectinesterase |
| PLN02933 | PLN02933 | 9.78e-10 | 33 | 310 | 213 | 468 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02488 | PLN02488 | 1.34e-09 | 35 | 303 | 194 | 444 | probable pectinesterase/pectinesterase inhibitor |
| pfam01095 | Pectinesterase | 1.01e-08 | 39 | 313 | 1 | 260 | Pectinesterase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT65238.1 | 1.08e-153 | 33 | 434 | 661 | 1073 |
| QCD41145.1 | 5.52e-151 | 33 | 436 | 1033 | 1438 |
| QCP72097.1 | 3.04e-132 | 35 | 437 | 21 | 428 |
| QCD38407.1 | 3.04e-132 | 35 | 437 | 21 | 428 |
| AEF12641.1 | 2.47e-117 | 33 | 431 | 1037 | 1442 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 1.76e-12 | 29 | 303 | 22 | 308 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
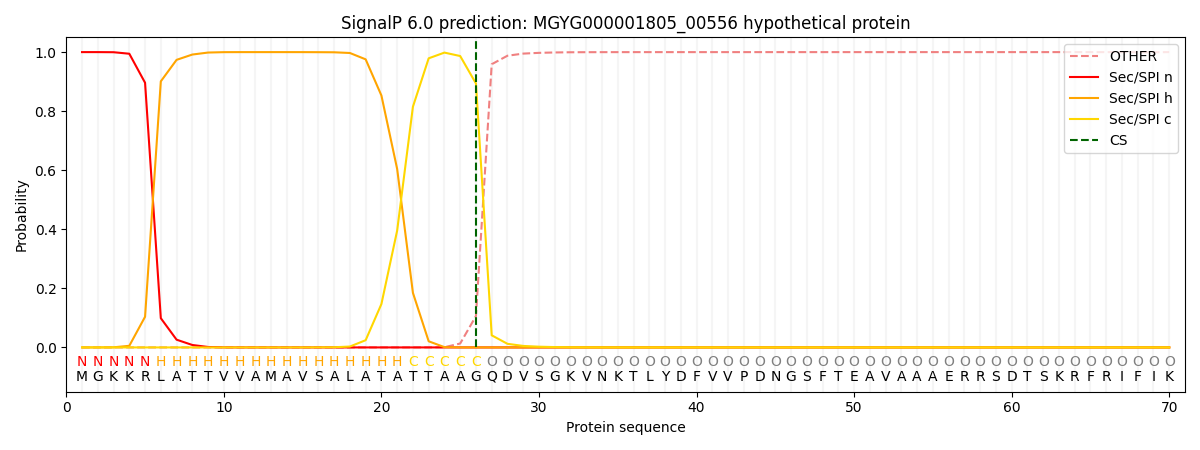
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000632 | 0.998370 | 0.000234 | 0.000282 | 0.000234 | 0.000209 |
