You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001805_01140
You are here: Home > Sequence: MGYG000001805_01140
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000001805_01140 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7410; End: 8939 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 205 | 483 | 1.2e-44 | 0.9479166666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02773 | PLN02773 | 3.76e-17 | 205 | 498 | 7 | 296 | pectinesterase |
| COG4677 | PemB | 9.66e-17 | 199 | 502 | 77 | 405 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02432 | PLN02432 | 6.56e-16 | 194 | 502 | 1 | 285 | putative pectinesterase |
| PLN02304 | PLN02304 | 5.31e-15 | 201 | 489 | 73 | 363 | probable pectinesterase |
| pfam01095 | Pectinesterase | 3.67e-13 | 206 | 473 | 3 | 271 | Pectinesterase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT59621.1 | 1.83e-289 | 14 | 506 | 8 | 500 |
| QQY40500.1 | 1.83e-289 | 14 | 506 | 8 | 500 |
| QQY44776.1 | 1.83e-289 | 14 | 506 | 8 | 500 |
| ABR39479.1 | 2.56e-289 | 14 | 506 | 17 | 509 |
| AII65147.1 | 4.29e-288 | 14 | 506 | 8 | 500 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O23038 | 1.12e-14 | 177 | 508 | 63 | 393 | Probable pectinesterase 8 OS=Arabidopsis thaliana OX=3702 GN=PME8 PE=2 SV=2 |
| Q9LVQ0 | 8.15e-14 | 201 | 480 | 3 | 282 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| Q9FM79 | 5.83e-10 | 184 | 366 | 58 | 246 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| Q9ZQA3 | 1.53e-09 | 205 | 368 | 91 | 250 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
| Q9SIJ9 | 8.93e-09 | 196 | 492 | 41 | 325 | Putative pectinesterase 11 OS=Arabidopsis thaliana OX=3702 GN=PME11 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
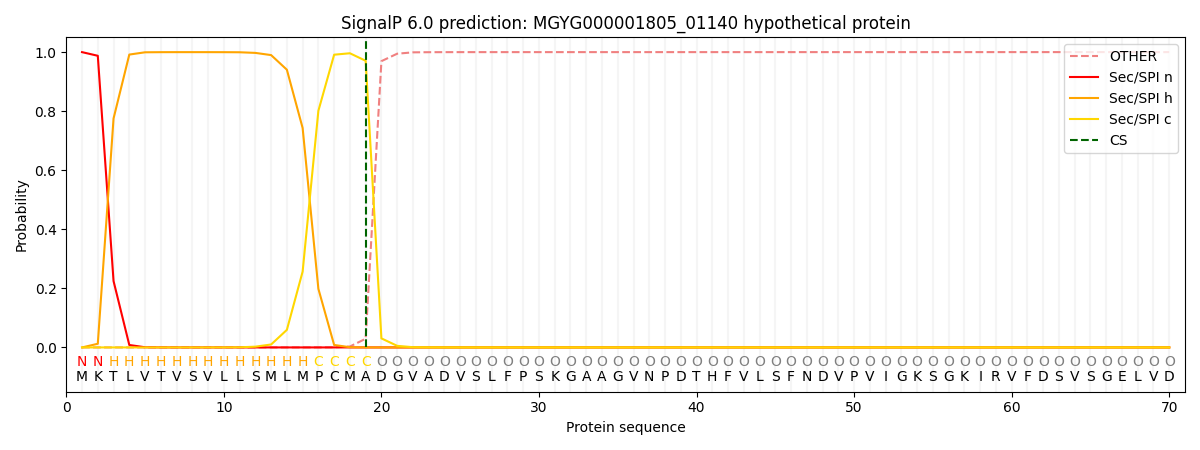
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000347 | 0.998508 | 0.000564 | 0.000195 | 0.000179 | 0.000164 |
