You are browsing environment: HUMAN GUT
MGYG000001807_00248
Basic Information
help
Species
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides;
CAZyme ID
MGYG000001807_00248
CAZy Family
GH5
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001807
2264331
MAG
Denmark
Europe
Gene Location
Start: 16174;
End: 18135
Strand: +
No EC number prediction in MGYG000001807_00248.
Family
Start
End
Evalue
family coverage
GH5
67
343
4e-87
0.9884169884169884
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam00150
Cellulase
2.56e-12
61
300
2
207
Cellulase (glycosyl hydrolase family 5).
more
pfam02368
Big_2
2.52e-04
456
513
26
77
Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins.
more
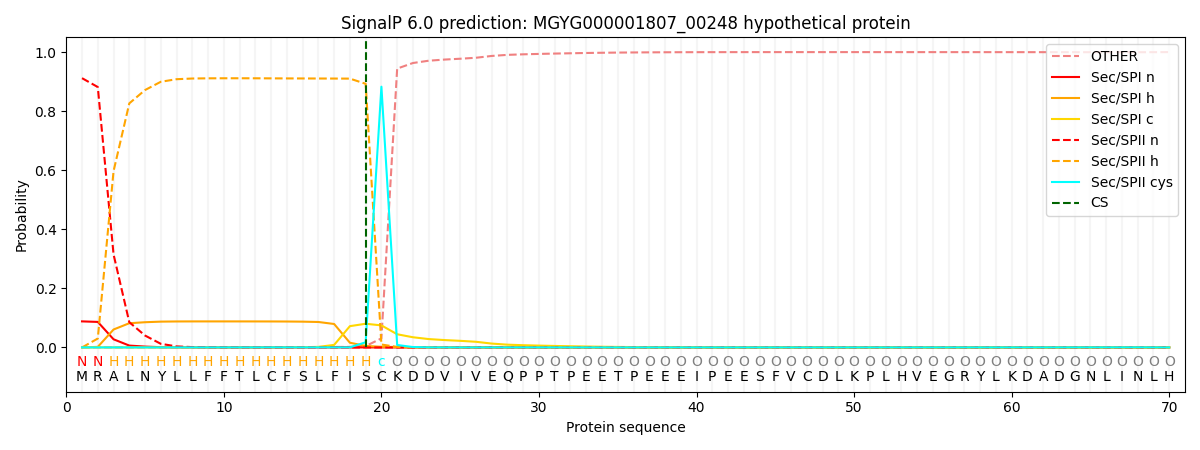
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000329
0.085991
0.913557
0.000045
0.000049
0.000043
There is no transmembrane helices in MGYG000001807_00248.