You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001810_01760
You are here: Home > Sequence: MGYG000001810_01760
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
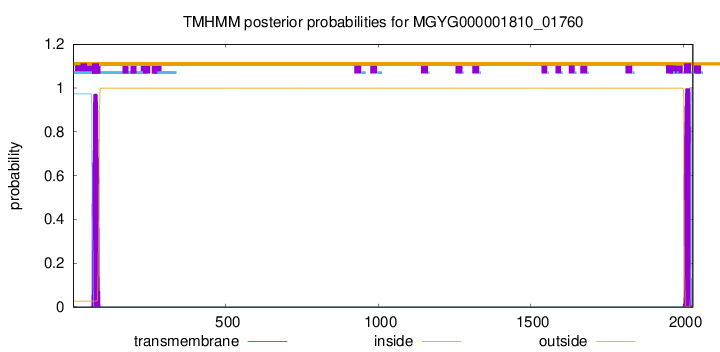
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp002314255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp002314255 | |||||||||||
| CAZyme ID | MGYG000001810_01760 | |||||||||||
| CAZy Family | GH101 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49419; End: 55517 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH101 | 570 | 1099 | 2.7e-85 | 0.7906647807637907 |
| CBM6 | 1605 | 1737 | 1.7e-23 | 0.855072463768116 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12905 | Glyco_hydro_101 | 3.57e-61 | 809 | 1072 | 1 | 269 | Endo-alpha-N-acetylgalactosaminidase. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by the S. pneumoniae protein Endo-alpha-N-acetylgalactosaminidase, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. |
| cd14244 | GH_101_like | 4.73e-61 | 824 | 1096 | 2 | 295 | Endo-a-N-acetylgalactosaminidase and related glcyosyl hydrolases. This family contains the enzymatically active domain of cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins (EC:3.2.1.97). It has been classified as glycosyl hydrolase family 101 in the Cazy resource. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae and other commensal human bacteria is largely determined by their ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. |
| pfam18080 | Gal_mutarotas_3 | 4.15e-35 | 566 | 808 | 1 | 243 | Galactose mutarotase-like fold domain. This domain is found in endo-alpha-N-acetylgalactosaminidase present in Streptococcus pneumoniae. Endo-alpha-N-acetylgalactosaminidase is a cell surface-anchored glycoside hydrolase involved in the breakdown of mucin type O-linked glycans. The domain, known as domain 2, exhibits strong structural similarlity to the galactose mutarotase-like fold but lacks the active site residues. Domains, found in a number of glycoside hydrolases, structurally similar to domain 2 confer stability to the multidomain architectures. |
| cd04084 | CBM6_xylanase-like | 1.65e-22 | 1602 | 1736 | 1 | 123 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
| pfam17451 | Glyco_hyd_101C | 1.18e-20 | 1084 | 1184 | 3 | 111 | Glycosyl hydrolase 101 beta sandwich domain. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by a S. pneumoniae protein, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. This domain represents C-terminal the beta sandwich domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATD58436.1 | 1.77e-223 | 112 | 1962 | 234 | 2123 |
| ATD54117.1 | 1.77e-223 | 112 | 1962 | 234 | 2123 |
| QBJ76357.1 | 1.77e-223 | 112 | 1962 | 234 | 2123 |
| SLK22612.1 | 1.77e-223 | 112 | 1962 | 234 | 2123 |
| BBK22105.1 | 1.19e-167 | 132 | 1586 | 67 | 1538 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6QEP_A | 4.31e-81 | 571 | 1460 | 17 | 1027 | EngBFDARPin Fusion 4b H14 [Bifidobacterium longum] |
| 6QFK_A | 4.39e-81 | 571 | 1460 | 17 | 1027 | EngBFDARPin Fusion 4b G10 [Bifidobacterium longum] |
| 6QEV_B | 4.39e-81 | 571 | 1460 | 17 | 1027 | EngBFDARPin Fusion 4b B6 [Bifidobacterium longum] |
| 6SH9_B | 4.39e-81 | 571 | 1460 | 17 | 1027 | EngBFDARPin Fusion 4b D12 [Bifidobacterium longum subsp. longum JCM 1217] |
| 2ZXQ_A | 4.98e-81 | 571 | 1460 | 32 | 1042 | Crystalstructure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A9WNA0 | 3.95e-76 | 571 | 1460 | 57 | 962 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
| Q2MGH6 | 4.46e-74 | 557 | 1587 | 323 | 1485 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0368 PE=1 SV=1 |
| Q8DR60 | 1.55e-72 | 557 | 1587 | 323 | 1485 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=spr0328 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
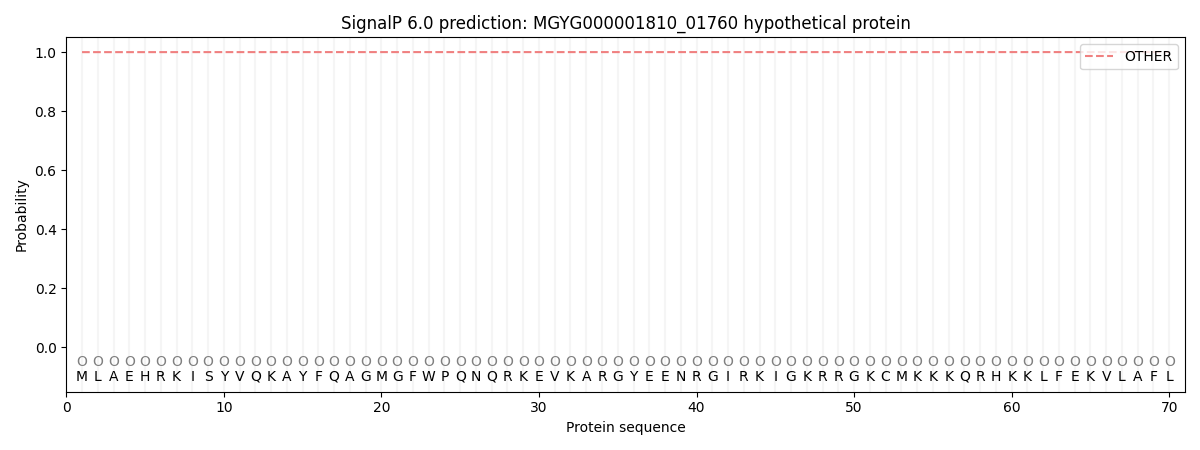
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999897 | 0.000089 | 0.000031 | 0.000001 | 0.000000 | 0.000008 |