You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001810_02448
You are here: Home > Sequence: MGYG000001810_02448
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp002314255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp002314255 | |||||||||||
| CAZyme ID | MGYG000001810_02448 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6320; End: 11755 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 61 | 613 | 4.1e-180 | 0.9507299270072993 |
| CBM51 | 1355 | 1509 | 2e-23 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 2.16e-22 | 1353 | 1510 | 2 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 2.65e-21 | 1354 | 1510 | 5 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam07081 | DUF1349 | 1.65e-08 | 894 | 1031 | 30 | 167 | Protein of unknown function (DUF1349). This family consists of several hypothetical bacterial proteins but contains one sequence from Saccharomyces cerevisiae. Members of this family are typically around 200 residues in length. The function of this family is unknown. |
| pfam02368 | Big_2 | 1.90e-07 | 762 | 845 | 1 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam13229 | Beta_helix | 1.47e-06 | 487 | 629 | 10 | 153 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBA55548.1 | 1.23e-232 | 52 | 851 | 17 | 841 |
| VEG17902.1 | 1.84e-231 | 20 | 992 | 12 | 964 |
| BAQ98494.1 | 2.65e-231 | 52 | 992 | 17 | 941 |
| AFL04872.1 | 2.65e-231 | 52 | 851 | 17 | 841 |
| QRI58269.1 | 3.05e-231 | 52 | 851 | 22 | 846 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JW4_A | 2.10e-48 | 57 | 611 | 24 | 568 | Crystalstructure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta],7JW4_B Crystal structure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta] |
| 7JWF_A | 9.20e-48 | 57 | 611 | 24 | 568 | Crystalstructure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_B Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_C Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_D Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta] |
| 7JRM_A | 5.08e-16 | 1333 | 1608 | 63 | 301 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JRL_A | 5.74e-16 | 1333 | 1608 | 85 | 323 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 2VMG_A | 1.41e-14 | 1338 | 1510 | 1 | 157 | Thestructure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1V8K7 | 2.92e-161 | 59 | 753 | 33 | 724 | Alpha-1,3-galactosidase A OS=Streptacidiphilus griseoplanus OX=66896 GN=glaA PE=1 SV=1 |
| Q826C5 | 1.32e-148 | 46 | 672 | 24 | 622 | Alpha-1,3-galactosidase A OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=glaA PE=3 SV=1 |
| Q8A2Z5 | 8.15e-90 | 99 | 610 | 46 | 533 | Alpha-1,3-galactosidase A OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=glaA PE=3 SV=1 |
| Q64MU6 | 4.09e-82 | 60 | 683 | 25 | 601 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=glaA PE=3 SV=1 |
| Q5L7M8 | 2.53e-81 | 60 | 683 | 25 | 601 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=glaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
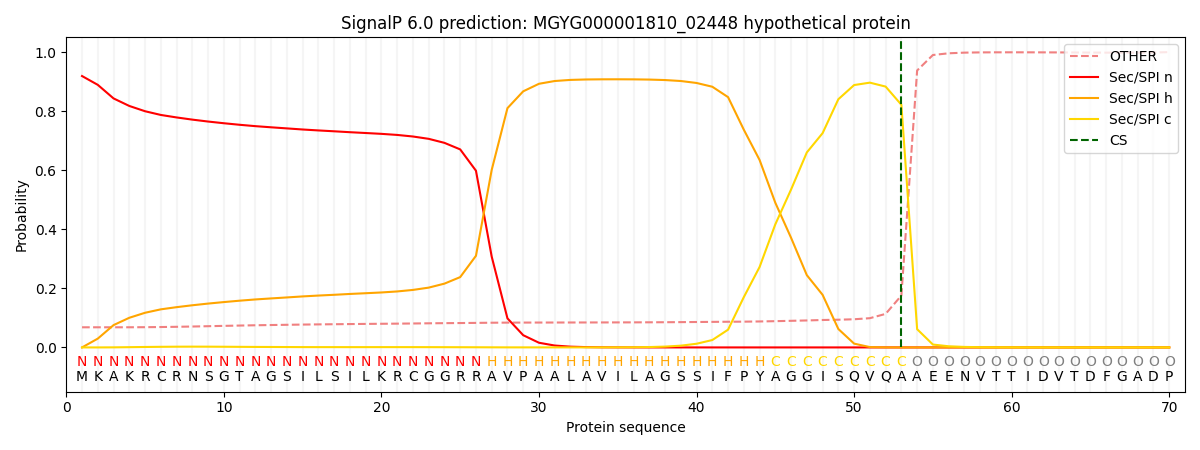
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.073360 | 0.910172 | 0.011418 | 0.003838 | 0.000820 | 0.000378 |

