You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001833_00363
You are here: Home > Sequence: MGYG000001833_00363
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
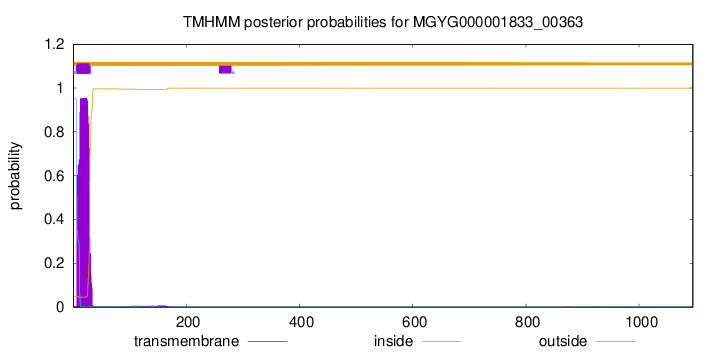
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; UMGS1441; | |||||||||||
| CAZyme ID | MGYG000001833_00363 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21510; End: 24797 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 635 | 985 | 1.3e-48 | 0.9583333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02682 | PLN02682 | 1.51e-23 | 627 | 991 | 68 | 368 | pectinesterase family protein |
| PLN02773 | PLN02773 | 2.22e-22 | 638 | 936 | 17 | 246 | pectinesterase |
| PLN02432 | PLN02432 | 7.14e-21 | 638 | 990 | 23 | 292 | putative pectinesterase |
| PLN02217 | PLN02217 | 1.75e-16 | 638 | 991 | 262 | 551 | probable pectinesterase/pectinesterase inhibitor |
| PLN02497 | PLN02497 | 8.79e-16 | 638 | 955 | 44 | 288 | probable pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAV09174.1 | 6.02e-142 | 182 | 1009 | 42 | 891 |
| ADL52345.1 | 4.35e-123 | 182 | 997 | 51 | 874 |
| BAV13168.1 | 4.73e-123 | 182 | 997 | 67 | 890 |
| ADD61757.1 | 6.94e-121 | 182 | 1011 | 23 | 846 |
| QSI02582.1 | 1.02e-114 | 182 | 1006 | 29 | 841 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A2QK82 | 2.34e-20 | 615 | 994 | 24 | 327 | Probable pectinesterase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pmeA PE=3 SV=1 |
| B0Y9F9 | 3.27e-19 | 616 | 940 | 22 | 274 | Probable pectinesterase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pmeA PE=3 SV=1 |
| Q4WBT5 | 3.27e-19 | 616 | 940 | 22 | 274 | Probable pectinesterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=pmeA PE=3 SV=1 |
| A1DBT4 | 5.11e-17 | 616 | 940 | 22 | 274 | Probable pectinesterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pmeA PE=3 SV=1 |
| Q12535 | 1.84e-16 | 782 | 994 | 110 | 330 | Pectinesterase OS=Aspergillus aculeatus OX=5053 GN=pme1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
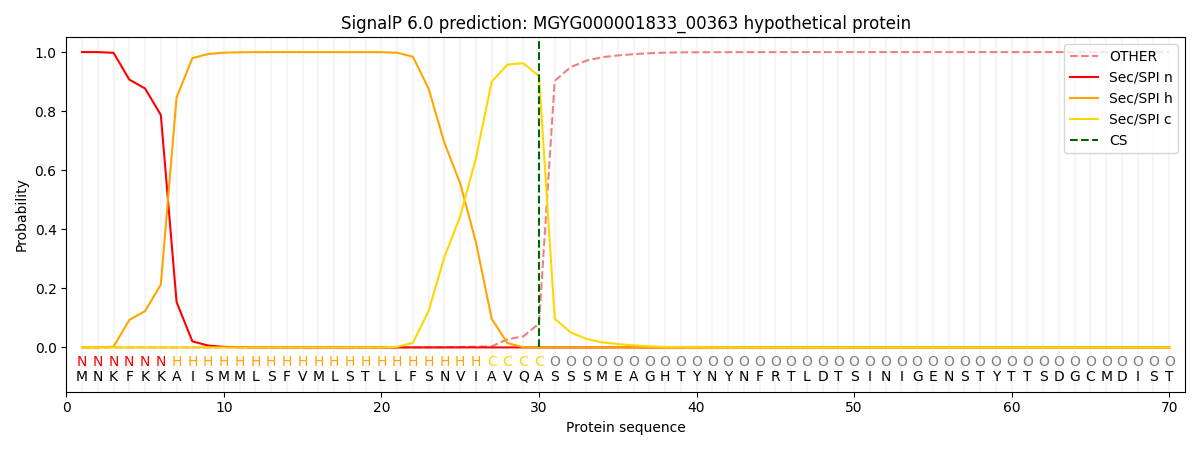
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000316 | 0.999006 | 0.000165 | 0.000182 | 0.000164 | 0.000148 |