You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001835_00480
You are here: Home > Sequence: MGYG000001835_00480
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
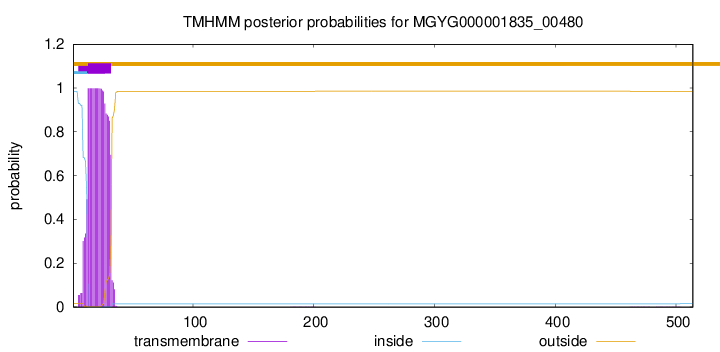
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp000432735 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp000432735 | |||||||||||
| CAZyme ID | MGYG000001835_00480 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 51154; End: 52698 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 436 | 513 | 3.4e-20 | 0.4350282485875706 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00912 | Transgly | 8.00e-23 | 442 | 513 | 17 | 88 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| PRK00056 | mtgA | 9.55e-22 | 441 | 514 | 63 | 137 | monofunctional biosynthetic peptidoglycan transglycosylase; Provisional |
| COG0744 | MrcB | 1.44e-18 | 440 | 513 | 78 | 152 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 1.32e-11 | 440 | 513 | 69 | 142 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| TIGR02073 | PBP_1c | 3.85e-09 | 442 | 514 | 37 | 109 | penicillin-binding protein 1C. This subfamily of the penicillin binding proteins includes the member from E. coli designated penicillin-binding protein 1C. Members have both transglycosylase and transpeptidase domains and are involved in forming cross-links in the late stages of peptidoglycan biosynthesis. All members of this subfamily are presumed to have the same basic function. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY37813.1 | 1.25e-209 | 1 | 514 | 1 | 513 |
| QUT79856.1 | 8.73e-162 | 2 | 514 | 6 | 513 |
| QDM09851.1 | 8.73e-162 | 2 | 514 | 6 | 513 |
| QNL39495.1 | 1.23e-161 | 2 | 514 | 6 | 513 |
| QGT72117.1 | 3.50e-161 | 2 | 514 | 6 | 513 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NB6_A | 2.72e-10 | 433 | 514 | 15 | 96 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
| 2OQO_A | 9.13e-10 | 433 | 514 | 15 | 96 | Crystalstructure of a peptidoglycan glycosyltransferase from a class A PBP: insight into bacterial cell wall synthesis [Aquifex aeolicus VF5],3D3H_A Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A [Aquifex aeolicus],3NB7_A Crystal structure of Aquifex Aeolicus Peptidoglycan Glycosyltransferase in complex with Decarboxylated Neryl Moenomycin [Aquifex aeolicus] |
| 3FWL_A | 3.64e-07 | 423 | 512 | 134 | 234 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli] |
| 5FGZ_A | 3.64e-07 | 423 | 512 | 130 | 230 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
| 3VMA_A | 3.67e-07 | 423 | 512 | 151 | 251 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2YBM4 | 1.16e-10 | 442 | 514 | 65 | 137 | Biosynthetic peptidoglycan transglycosylase OS=Nitrosospira multiformis (strain ATCC 25196 / NCIMB 11849 / C 71) OX=323848 GN=mtgA PE=3 SV=1 |
| Q7NS41 | 2.06e-10 | 442 | 514 | 63 | 135 | Biosynthetic peptidoglycan transglycosylase OS=Chromobacterium violaceum (strain ATCC 12472 / DSM 30191 / JCM 1249 / NBRC 12614 / NCIMB 9131 / NCTC 9757) OX=243365 GN=mtgA PE=3 SV=1 |
| C1D8R6 | 3.76e-10 | 420 | 514 | 38 | 135 | Biosynthetic peptidoglycan transglycosylase OS=Laribacter hongkongensis (strain HLHK9) OX=557598 GN=mtgA PE=3 SV=1 |
| Q3SFZ8 | 5.37e-10 | 442 | 512 | 66 | 136 | Biosynthetic peptidoglycan transglycosylase OS=Thiobacillus denitrificans (strain ATCC 25259) OX=292415 GN=mtgA PE=3 SV=1 |
| Q478U7 | 7.15e-10 | 442 | 514 | 66 | 138 | Biosynthetic peptidoglycan transglycosylase OS=Dechloromonas aromatica (strain RCB) OX=159087 GN=mtgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
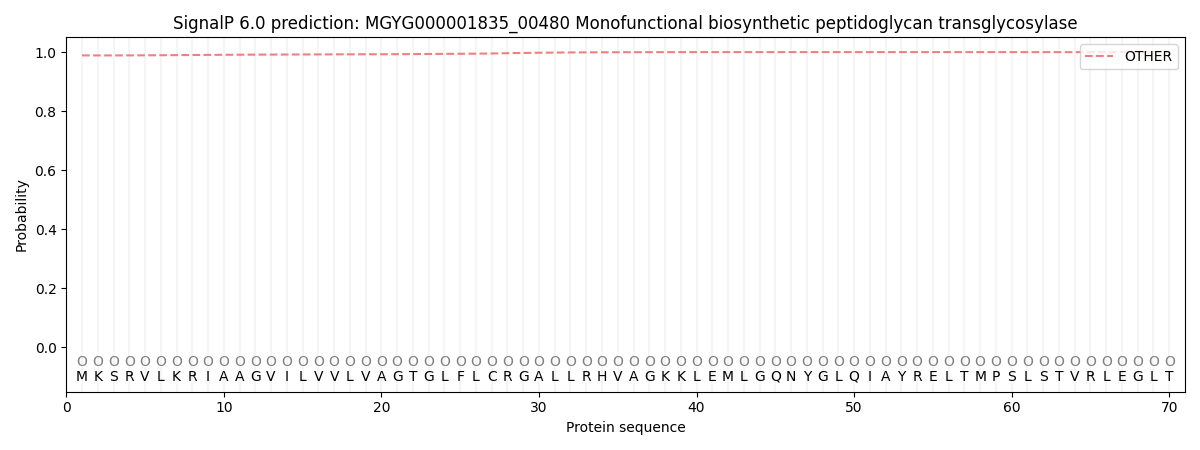
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.989175 | 0.006967 | 0.001713 | 0.000042 | 0.000025 | 0.002097 |