You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001840_00559
You are here: Home > Sequence: MGYG000001840_00559
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
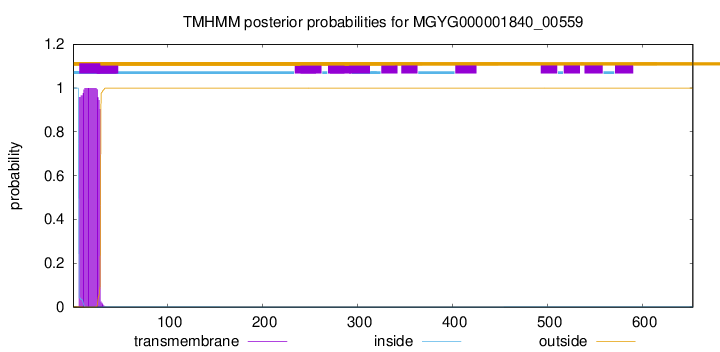
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; UMGS2016; | |||||||||||
| CAZyme ID | MGYG000001840_00559 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 2D | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27698; End: 29659 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 56 | 221 | 2.6e-64 | 0.9378531073446328 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 0.0 | 21 | 615 | 27 | 623 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 2.14e-131 | 59 | 653 | 67 | 781 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 8.43e-82 | 56 | 222 | 10 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| PRK09506 | mrcB | 4.32e-72 | 72 | 567 | 224 | 724 | bifunctional glycosyl transferase/transpeptidase; Reviewed |
| COG4953 | PbpC | 2.20e-71 | 23 | 567 | 16 | 538 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXI38555.1 | 4.75e-187 | 56 | 636 | 71 | 652 |
| ASS89543.1 | 4.75e-187 | 56 | 636 | 71 | 652 |
| QNU38784.1 | 1.26e-186 | 24 | 636 | 36 | 653 |
| BBU41270.1 | 1.90e-186 | 56 | 636 | 71 | 652 |
| QXJ38547.1 | 2.52e-186 | 24 | 636 | 36 | 653 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DWK_A | 3.87e-67 | 45 | 564 | 10 | 557 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 2OLU_A | 1.10e-62 | 29 | 564 | 9 | 566 | StructuralInsight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme [Staphylococcus aureus],2OLV_A Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus],2OLV_B Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus] |
| 5FGZ_A | 4.12e-59 | 56 | 568 | 153 | 672 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
| 3VMA_A | 5.39e-59 | 56 | 568 | 174 | 693 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli K-12] |
| 3FWL_A | 5.01e-57 | 56 | 568 | 157 | 676 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P70997 | 6.52e-164 | 54 | 636 | 69 | 653 | Penicillin-binding protein 2D OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpG PE=2 SV=3 |
| O66874 | 2.79e-82 | 45 | 636 | 47 | 712 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
| P38050 | 1.71e-75 | 15 | 565 | 24 | 571 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
| P39793 | 1.55e-68 | 34 | 636 | 72 | 691 | Penicillin-binding protein 1A/1B OS=Bacillus subtilis (strain 168) OX=224308 GN=ponA PE=1 SV=1 |
| Q9KUC0 | 4.52e-68 | 58 | 568 | 179 | 693 | Penicillin-binding protein 1B OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=mrcB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
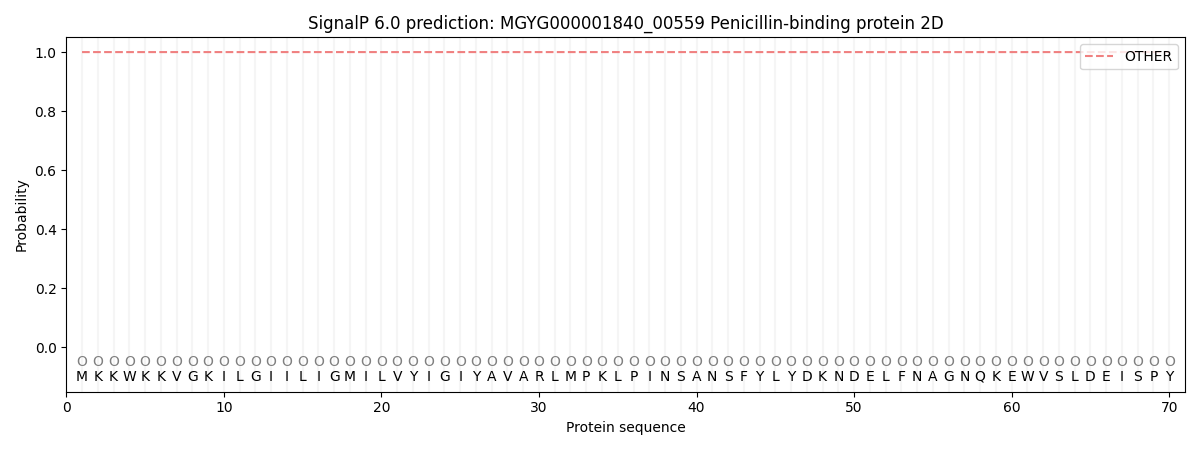
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000045 | 0.000003 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |