You are browsing environment: HUMAN GUT
MGYG000001850_01389
Basic Information
help
Species
Lineage
Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; QALW01; UMGS1322;
CAZyme ID
MGYG000001850_01389
CAZy Family
GH140
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
623
70865.94
4.2091
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001850
2350438
MAG
Denmark
Europe
Gene Location
Start: 19223;
End: 21094
Strand: -
No EC number prediction in MGYG000001850_01389.
Family
Start
End
Evalue
family coverage
GH140
432
618
2.6e-18
0.3737864077669903
GH140
164
353
1e-16
0.4320388349514563
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam13204
DUF4038
2.99e-36
164
504
1
318
Protein of unknown function (DUF4038). A family of putative cellulases.
more
pfam16586
DUF5060
5.87e-10
60
130
3
69
Domain of unknown function (DUF5060). This is the N-terminal domain of a putative glycoside hydrolase, DUF4038. It is found in a number of different bacterial orders.
more
pfam12904
Collagen_bind_2
1.21e-04
543
618
20
89
Putative collagen-binding domain of a collagenase. This domain is likely to be the collagen-binding domain of a family of bacterial collagenase enzymes. It is the C-terminal part of the Structure 3kzs (information derived from TOPSAN).
more
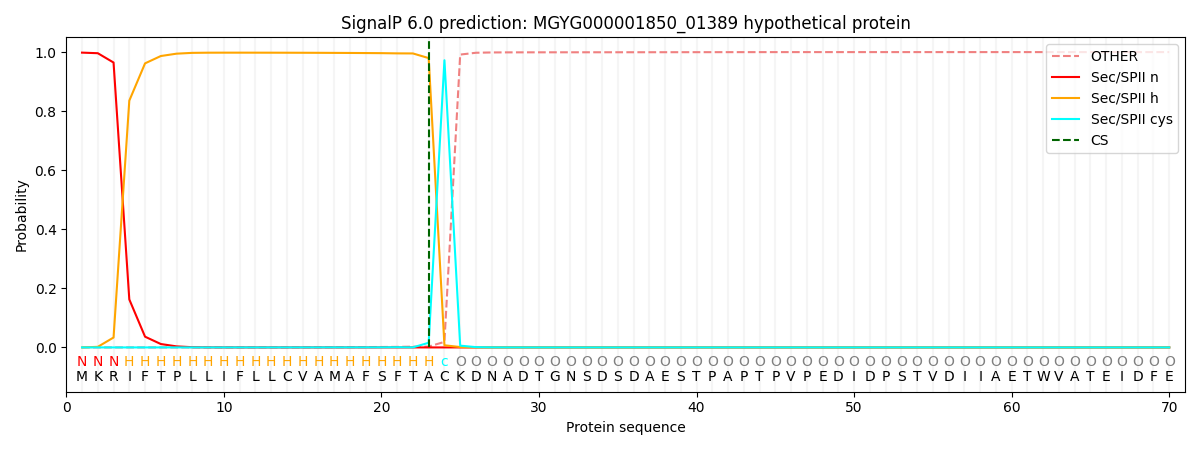
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000018
0.001690
0.998307
0.000002
0.000003
0.000002
There is no transmembrane helices in MGYG000001850_01389.