You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001850_01678
You are here: Home > Sequence: MGYG000001850_01678
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; QALW01; UMGS1322; | |||||||||||
| CAZyme ID | MGYG000001850_01678 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13821; End: 18647 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 1149 | 1381 | 1.1e-46 | 0.9237668161434978 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16192 | PMT_4TMC | 3.81e-21 | 1399 | 1602 | 7 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG1928 | PMT1 | 2.24e-16 | 1399 | 1606 | 505 | 695 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG5542 | COG5542 | 1.88e-15 | 282 | 629 | 71 | 394 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| COG1928 | PMT1 | 3.93e-10 | 1151 | 1379 | 46 | 265 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| pfam13231 | PMT_2 | 5.19e-10 | 305 | 437 | 1 | 130 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CQR52353.1 | 5.68e-180 | 30 | 1606 | 30 | 1275 |
| AIQ39188.1 | 3.38e-175 | 30 | 1606 | 30 | 1279 |
| AIQ56104.1 | 4.70e-175 | 30 | 1606 | 30 | 1279 |
| AIQ27397.1 | 1.26e-174 | 30 | 1606 | 30 | 1279 |
| QUL55595.1 | 2.40e-172 | 30 | 1606 | 30 | 1278 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| L8F4Z2 | 2.81e-10 | 1155 | 1455 | 67 | 394 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
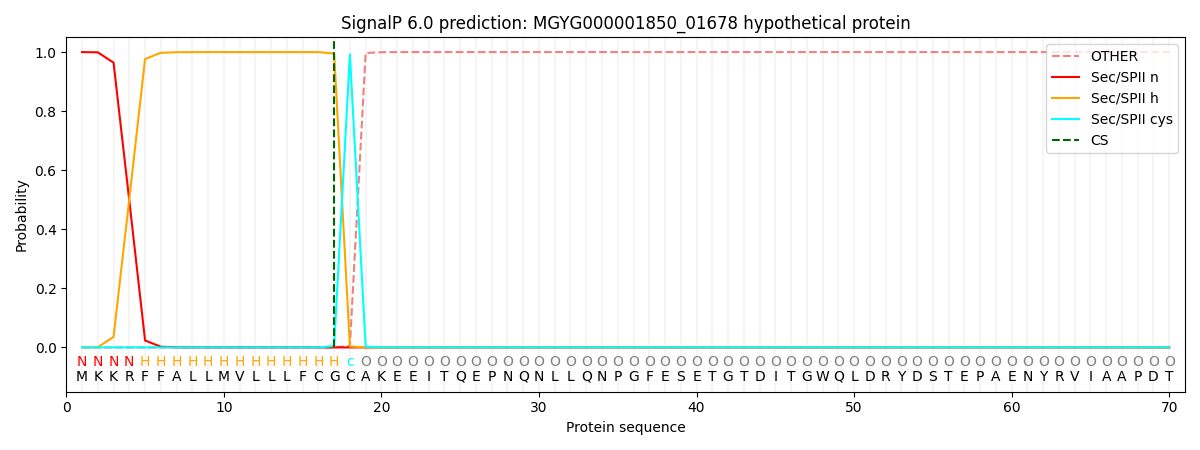
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000034 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
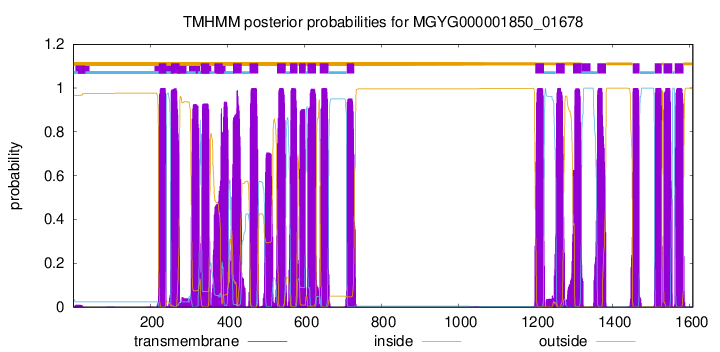
| start | end |
|---|---|
| 222 | 241 |
| 254 | 276 |
| 331 | 353 |
| 366 | 388 |
| 415 | 437 |
| 458 | 480 |
| 529 | 551 |
| 563 | 582 |
| 586 | 603 |
| 608 | 630 |
| 640 | 662 |
| 710 | 729 |
| 1199 | 1221 |
| 1253 | 1275 |
| 1297 | 1319 |
| 1360 | 1382 |
| 1452 | 1469 |
| 1509 | 1527 |
| 1532 | 1554 |
| 1561 | 1583 |
