You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001850_02037
You are here: Home > Sequence: MGYG000001850_02037
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; QALW01; UMGS1322; | |||||||||||
| CAZyme ID | MGYG000001850_02037 | |||||||||||
| CAZy Family | GH140 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 316; End: 2046 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH140 | 144 | 571 | 6.2e-34 | 0.9951456310679612 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13204 | DUF4038 | 2.64e-32 | 150 | 473 | 1 | 320 | Protein of unknown function (DUF4038). A family of putative cellulases. |
| pfam16586 | DUF5060 | 3.95e-24 | 49 | 117 | 2 | 70 | Domain of unknown function (DUF5060). This is the N-terminal domain of a putative glycoside hydrolase, DUF4038. It is found in a number of different bacterial orders. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCW44877.1 | 1.24e-141 | 61 | 575 | 94 | 592 |
| QTH45264.1 | 4.60e-107 | 50 | 575 | 41 | 569 |
| QJD82871.1 | 3.72e-105 | 50 | 575 | 41 | 569 |
| QNK56434.1 | 9.80e-101 | 46 | 573 | 34 | 565 |
| BBI36498.1 | 2.31e-99 | 46 | 576 | 35 | 569 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4N0R_A | 2.76e-11 | 63 | 359 | 9 | 307 | ChainA, putative glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],4N0R_B Chain B, putative glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
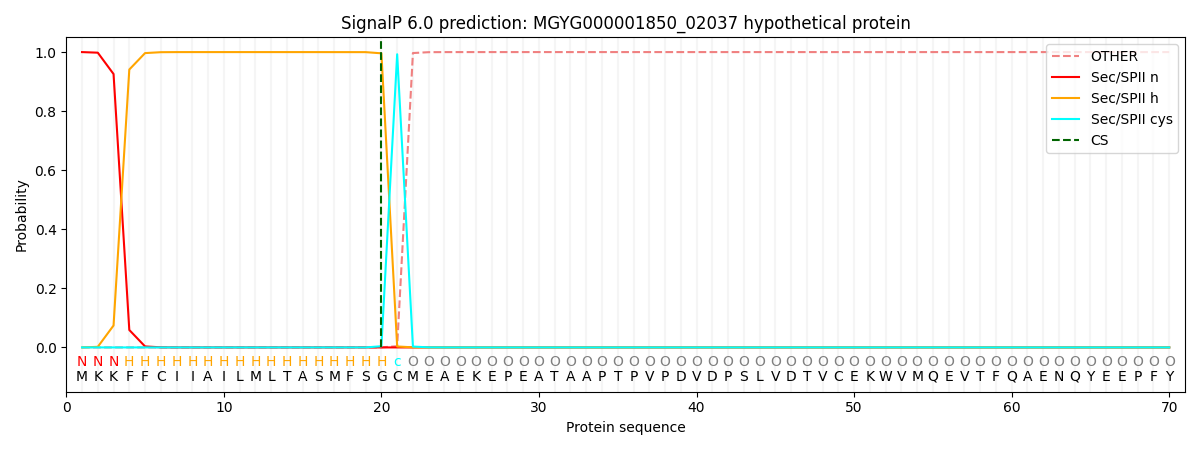
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000044 | 0.000000 | 0.000000 | 0.000000 |
