You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001854_00807
You are here: Home > Sequence: MGYG000001854_00807
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
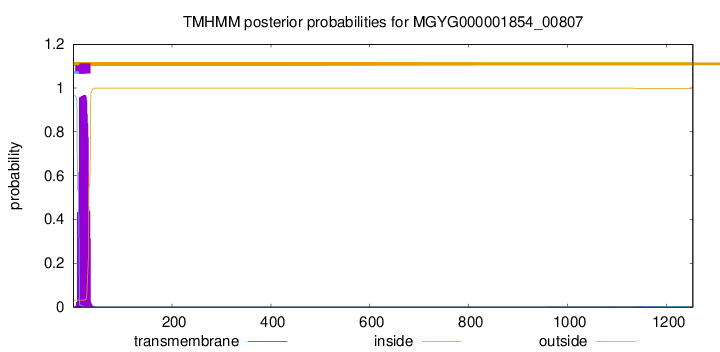
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; Borkfalkiaceae; UBA11940; | |||||||||||
| CAZyme ID | MGYG000001854_00807 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 39147; End: 42908 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 56 | 520 | 1.2e-98 | 0.9786666666666667 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 5.85e-04 | 148 | 285 | 2 | 114 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam05342 | Peptidase_M26_N | 0.002 | 871 | 956 | 143 | 226 | M26 IgA1-specific Metallo-endopeptidase N-terminal region. These peptidases, which cleave mammalian IgA, are found in Gram-positive bacteria. Often found associated with pfam00746, they may be attached to the cell wall. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAG59609.1 | 3.55e-75 | 61 | 529 | 410 | 825 |
| ADL51369.1 | 1.68e-73 | 61 | 529 | 834 | 1249 |
| ABX42257.1 | 1.58e-66 | 61 | 526 | 204 | 610 |
| AWB44904.1 | 3.59e-66 | 21 | 516 | 15 | 432 |
| AIQ18143.1 | 2.17e-65 | 61 | 368 | 2 | 283 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 5.90e-39 | 58 | 382 | 15 | 306 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 1.13e-13 | 57 | 374 | 2 | 325 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A6 | 2.65e-39 | 58 | 382 | 40 | 331 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 5.38e-38 | 58 | 382 | 40 | 331 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P22751 | 1.15e-19 | 58 | 368 | 389 | 639 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
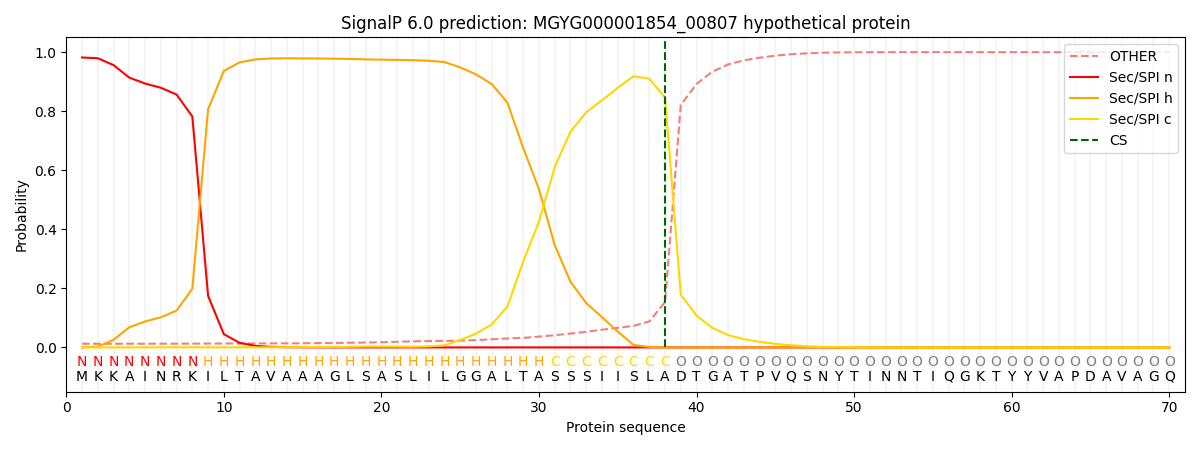
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.014106 | 0.978357 | 0.005516 | 0.001386 | 0.000329 | 0.000259 |