You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001854_01218
You are here: Home > Sequence: MGYG000001854_01218
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; Borkfalkiaceae; UBA11940; | |||||||||||
| CAZyme ID | MGYG000001854_01218 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8298; End: 11096 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 446 | 617 | 3.9e-45 | 0.8267326732673267 |
| CBM77 | 834 | 924 | 4.2e-20 | 0.8640776699029126 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 1.32e-56 | 315 | 735 | 2 | 345 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 1.01e-35 | 439 | 614 | 13 | 186 | Amb_all domain. |
| pfam18283 | CBM77 | 2.94e-24 | 816 | 926 | 1 | 108 | Carbohydrate binding module 77. This domain is the non-catalytic carbohydrate binding module 77 (CBM77) present in Ruminococcus flavefaciens. CBMs fulfil a critical targeting function in plant cell wall depolymerisation. In CBM77, a cluster of conserved basic residues (Lys1092, Lys1107 and Lys1162) confer calcium-independent recognition of homogalacturonan. |
| pfam00544 | Pec_lyase_C | 5.48e-22 | 456 | 614 | 42 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| NF035936 | agg_sub_LPXTH | 4.66e-09 | 10 | 203 | 20 | 189 | serine-rich aggregation substance UasX. Members of this protein family are repetitive, serine-rich surface proteins of the Firmicutes, found primarily in the genus Leuconostoc. The variant form of sortase signal, LPXTH, is replaced by LPXTG in members from some lineages, such as Weissella oryzae, and therefore recognizable. Some members of this family have the KxYKxGKxW type signal peptide as seen in the glycoprotein adhesin GspB, a substrate of the accessory Sec system for secretion. WOSG25_050600 from Weissella oryzae SG25 is identified in a publication as an unnamed aggregation substance, a conclusion supported by the sorting signals and composition reported here. We assign the gene symbol uasX (unnamed aggregation substance X) based on our evaluation of the family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL35105.1 | 2.77e-199 | 249 | 779 | 48 | 591 |
| ADU23076.1 | 1.02e-191 | 247 | 927 | 42 | 704 |
| QHJ07965.1 | 3.47e-184 | 252 | 924 | 18 | 664 |
| QDW19698.1 | 5.99e-184 | 255 | 769 | 22 | 554 |
| AXB56096.1 | 1.09e-183 | 252 | 769 | 19 | 554 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VMV_A | 5.15e-23 | 354 | 614 | 6 | 246 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 1AIR_A | 1.41e-18 | 393 | 727 | 15 | 339 | ChainA, PECTATE LYASE C [Dickeya chrysanthemi],1O88_A Chain A, Pectate Lyase C [Dickeya chrysanthemi],1O8D_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8E_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8F_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8G_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8H_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8I_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8J_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8K_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8L_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8M_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1PLU_A Chain A, Protein (pectate Lyase C) [Dickeya chrysanthemi],2PEC_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi] |
| 2EWE_A | 3.41e-18 | 393 | 727 | 15 | 339 | ChainA, Pectate lyase C [Dickeya chrysanthemi] |
| 1VBL_A | 6.58e-16 | 455 | 624 | 138 | 340 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 5AMV_A | 3.17e-13 | 449 | 612 | 127 | 323 | Structuralinsights into the loss of catalytic competence in pectate lyase at low pH [Bacillus subtilis],5X2I_A Polygalacturonate Lyase by Fusing with a Self-assembling Amphipathic Peptide [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1B6T1 | 2.00e-22 | 394 | 616 | 55 | 274 | Pectate trisaccharide-lyase OS=Bacillus sp. OX=1409 GN=pel PE=1 SV=1 |
| Q8GCB2 | 2.00e-22 | 394 | 616 | 55 | 274 | Pectate trisaccharide-lyase OS=Bacillus licheniformis OX=1402 GN=pelA PE=1 SV=1 |
| Q65DC2 | 2.00e-22 | 394 | 616 | 55 | 274 | Pectate trisaccharide-lyase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=BLi04129 PE=3 SV=1 |
| B0XT32 | 2.04e-18 | 456 | 640 | 97 | 282 | Probable pectate lyase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyA PE=3 SV=1 |
| Q4WIT0 | 2.04e-18 | 456 | 640 | 97 | 282 | Probable pectate lyase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
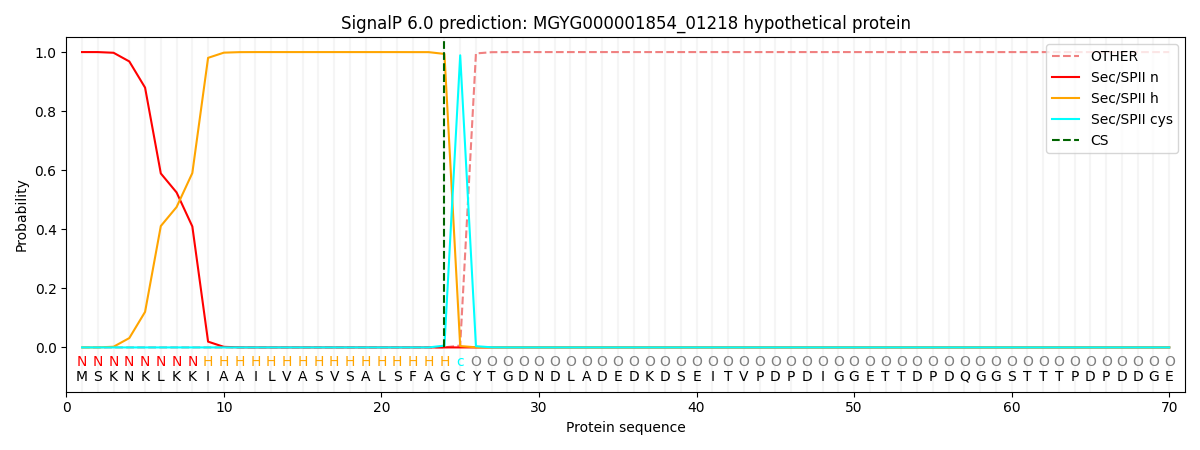
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000002 | 1.000062 | 0.000000 | 0.000000 | 0.000000 |
