You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001867_00057
You are here: Home > Sequence: MGYG000001867_00057
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
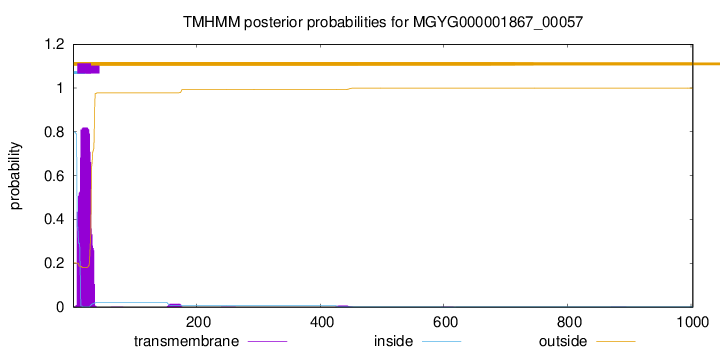
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900555715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900555715 | |||||||||||
| CAZyme ID | MGYG000001867_00057 | |||||||||||
| CAZy Family | GH115 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 31209; End: 34220 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH115 | 43 | 671 | 7.2e-214 | 0.8278335724533716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam15979 | Glyco_hydro_115 | 9.96e-160 | 196 | 537 | 2 | 333 | Glycosyl hydrolase family 115. Glyco_hydro_115 is a family of glycoside hydrolases likely to have the activity of xylan a-1,2-glucuronidase, EC:3.2.1.131, or a-(4-O-methyl)-glucuronidase EC:3.2.1.-. |
| pfam17829 | GH115_C | 3.06e-52 | 811 | 987 | 2 | 170 | Gylcosyl hydrolase family 115 C-terminal domain. This domain is found at the C-terminus of glycosyl hydrolase family 115 proteins. This domain has a beta-sandwich fold. |
| pfam05124 | S_layer_C | 0.001 | 54 | 165 | 91 | 220 | S-layer like family, C-terminal region. |
| cd14948 | BACON | 0.009 | 736 | 795 | 26 | 81 | Bacteroidetes-Associated Carbohydrate-binding (putative) Often N-terminal (BACON) domain. The BACON domain is found in diverse domain architectures and accociated with a wide variety of domains, including carbohydrate-active enzymes and proteases. It was named for its suggested function of carbohydrate binding; the latter was inferred from domain architectures, sequence conservation, and phyletic distribution. However, recent experimental data suggest that its primary function in Bacteroides ovatus endo-xyloglucanase BoGH5A is to distance the catalytic module from the cell surface and confer additional mobility to the catalytic domain for attack of the polysaccharide. No evidence for a direct role in carbohydrate binding could be found in that case. The large majority of BACON domains are found in Bacteroidetes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT41413.1 | 0.0 | 15 | 990 | 4 | 979 |
| QMW85894.1 | 0.0 | 15 | 990 | 4 | 979 |
| AAO78620.1 | 0.0 | 15 | 990 | 4 | 979 |
| QQA08355.1 | 0.0 | 15 | 990 | 4 | 979 |
| ALJ44299.1 | 0.0 | 15 | 990 | 4 | 979 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZMH_A | 3.42e-141 | 37 | 987 | 7 | 930 | Crystalstructure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40],4ZMH_B Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40] |
| 5BY3_A | 7.83e-124 | 64 | 660 | 39 | 611 | Anovel family GH115 4-O-Methyl-alpha-glucuronidase, BtGH115A, with specificity for decorated arabinogalactans [Bacteroides thetaiotaomicron VPI-5482] |
| 4C90_A | 1.26e-108 | 44 | 741 | 44 | 705 | Evidencethat GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C90_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_A Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus] |
| 6NPS_A | 6.43e-95 | 50 | 993 | 17 | 966 | Crystalstructure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112],6NPS_B Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112] |
| 7PUG_A | 1.45e-93 | 91 | 668 | 63 | 629 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
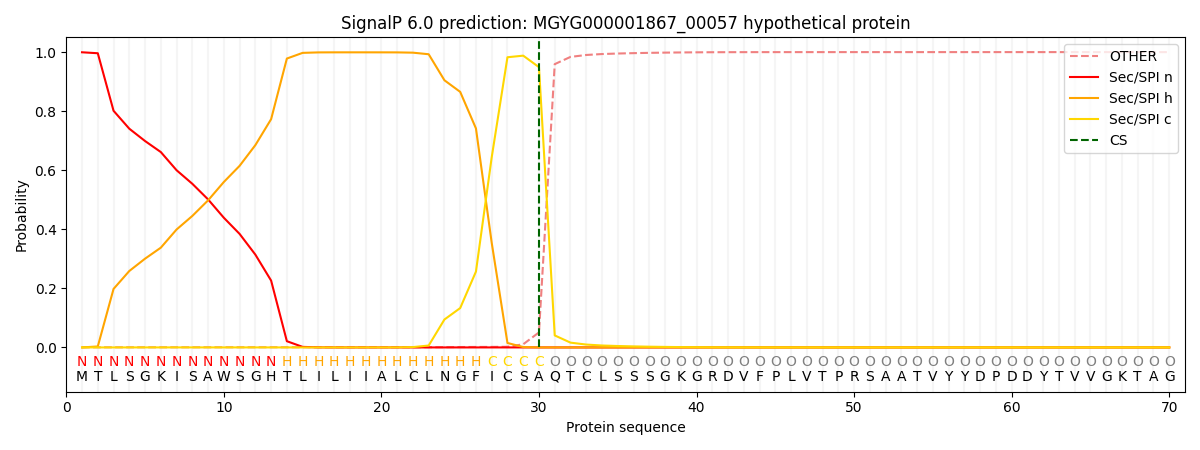
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000967 | 0.998108 | 0.000427 | 0.000165 | 0.000151 | 0.000151 |