You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001871_00958
You are here: Home > Sequence: MGYG000001871_00958
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; ; | |||||||||||
| CAZyme ID | MGYG000001871_00958 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 177542; End: 180331 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH128 | 244 | 466 | 4.5e-43 | 0.9598214285714286 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam11790 | Glyco_hydro_cc | 2.55e-35 | 246 | 466 | 19 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| pfam01229 | Glyco_hydro_39 | 2.30e-06 | 299 | 401 | 155 | 274 | Glycosyl hydrolases family 39. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB49856.1 | 7.01e-153 | 17 | 916 | 42 | 903 |
| AXY72824.1 | 4.88e-96 | 18 | 563 | 384 | 893 |
| QOG04727.1 | 1.19e-91 | 23 | 783 | 550 | 1285 |
| BAV05333.1 | 6.85e-91 | 24 | 589 | 540 | 1085 |
| AEV99551.1 | 2.71e-90 | 29 | 528 | 417 | 891 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6UAQ_A | 5.83e-22 | 247 | 444 | 47 | 236 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei],6UAR_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose [Amycolatopsis mediterranei] |
| 6UFL_A | 1.45e-21 | 247 | 444 | 47 | 236 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose [Amycolatopsis mediterranei],6UFZ_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei] |
| 6UAS_A | 3.60e-21 | 247 | 444 | 47 | 236 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei] |
| 6UAT_A | 3.60e-21 | 247 | 444 | 47 | 236 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei],6UAU_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose and laminaribiose [Amycolatopsis mediterranei] |
| 6UB0_A | 1.10e-13 | 246 | 443 | 24 | 209 | Crystalstructure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -2 and -1 subsites [Blastomyces gilchristii SLH14081] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
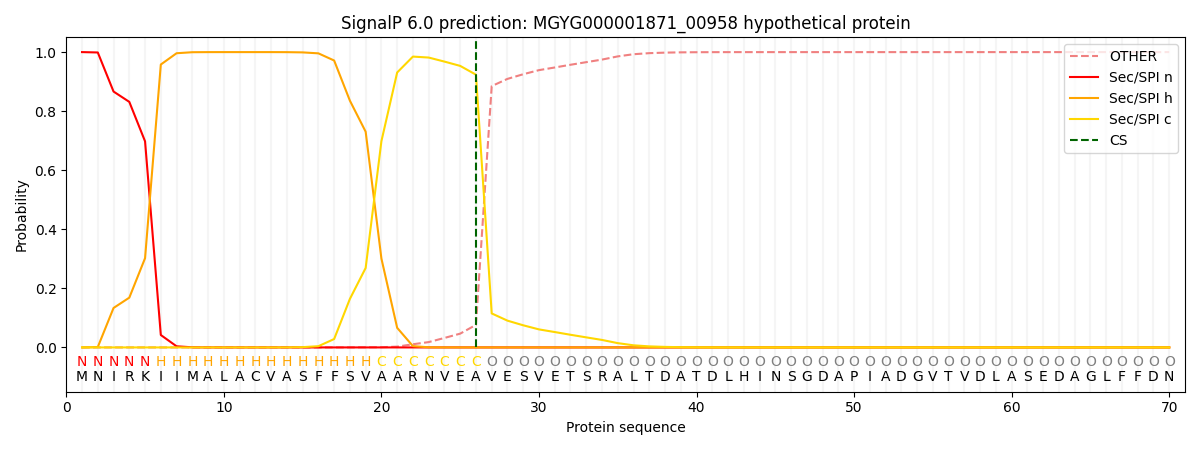
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000263 | 0.999017 | 0.000202 | 0.000178 | 0.000161 | 0.000139 |
