You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001873_00173
You are here: Home > Sequence: MGYG000001873_00173
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Marvinbryantia; | |||||||||||
| CAZyme ID | MGYG000001873_00173 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5438; End: 7864 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 261 | 482 | 6.2e-41 | 0.8287037037037037 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 7.60e-27 | 243 | 581 | 78 | 361 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK15098 | PRK15098 | 4.68e-19 | 263 | 804 | 127 | 648 | beta-glucosidase BglX. |
| pfam00933 | Glyco_hydro_3 | 1.12e-15 | 284 | 482 | 116 | 282 | Glycosyl hydrolase family 3 N terminal domain. |
| PLN03080 | PLN03080 | 2.10e-10 | 291 | 570 | 154 | 415 | Probable beta-xylosidase; Provisional |
| pfam01915 | Glyco_hydro_3_C | 1.39e-05 | 558 | 735 | 1 | 189 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIK51969.1 | 1.48e-169 | 34 | 804 | 16 | 783 |
| QIA42793.1 | 2.86e-169 | 35 | 804 | 19 | 784 |
| ATL89160.1 | 2.86e-169 | 35 | 804 | 19 | 784 |
| ATO98801.1 | 2.86e-169 | 35 | 804 | 19 | 784 |
| ARE65206.1 | 3.60e-139 | 51 | 804 | 22 | 770 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M6G_A | 2.51e-22 | 262 | 751 | 149 | 603 | Crystalstructure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea [Saccharopolyspora erythraea D] |
| 5YOT_A | 2.55e-21 | 270 | 734 | 130 | 582 | Isoprimeverose-producingenzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YOT_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_A Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40] |
| 7EAP_A | 4.43e-21 | 270 | 734 | 130 | 582 | ChainA, Fn3_like domain-containing protein [Aspergillus oryzae RIB40] |
| 5Z87_A | 1.81e-17 | 276 | 736 | 156 | 630 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 3USZ_A | 3.88e-16 | 262 | 585 | 130 | 425 | Crystalstructure of truncated exo-1,3/1,4-beta-glucanase (EXOP) from Pseudoalteromonas sp. BB1 [Pseudoalteromonas sp. BB1],3UT0_A Crystal structure of exo-1,3/1,4-beta-glucanase (EXOP) from Pseudoalteromonas sp. BB1 [Pseudoalteromonas sp. BB1],3UT0_B Crystal structure of exo-1,3/1,4-beta-glucanase (EXOP) from Pseudoalteromonas sp. BB1 [Pseudoalteromonas sp. BB1],3UT0_C Crystal structure of exo-1,3/1,4-beta-glucanase (EXOP) from Pseudoalteromonas sp. BB1 [Pseudoalteromonas sp. BB1],3UT0_D Crystal structure of exo-1,3/1,4-beta-glucanase (EXOP) from Pseudoalteromonas sp. BB1 [Pseudoalteromonas sp. BB1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 7.40e-42 | 64 | 737 | 33 | 625 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| Q2UFP8 | 3.99e-40 | 234 | 742 | 138 | 602 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| B8NGU6 | 9.29e-40 | 234 | 742 | 134 | 598 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| Q5BCC6 | 1.78e-34 | 234 | 751 | 126 | 597 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| T2KMH0 | 2.76e-16 | 270 | 575 | 104 | 388 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
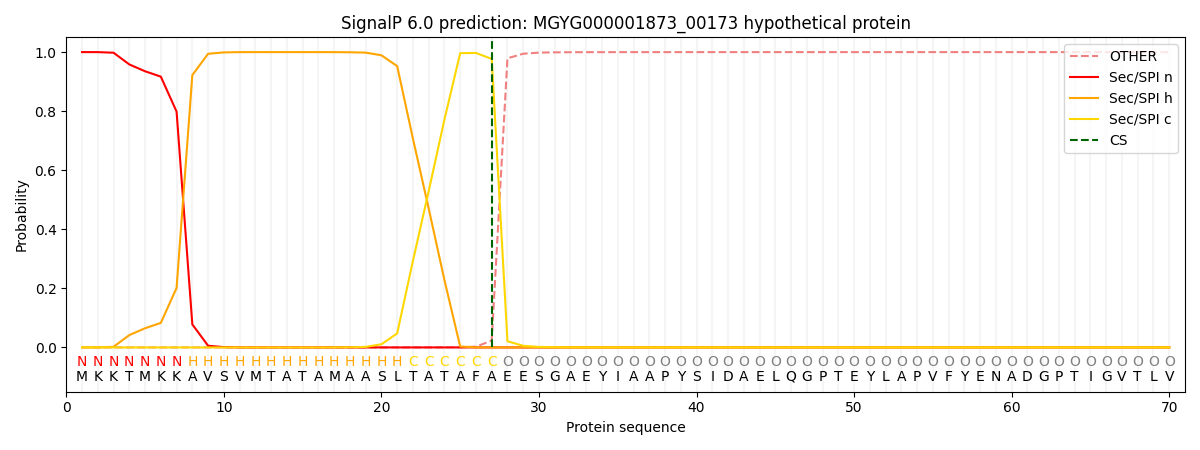
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000511 | 0.998288 | 0.000275 | 0.000346 | 0.000307 | 0.000239 |
