You are browsing environment: HUMAN GUT
MGYG000001878_00247
Basic Information
help
Species
Parabacteroides sp900548175
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900548175
CAZyme ID
MGYG000001878_00247
CAZy Family
GT83
CAZyme Description
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
CAZyme Property
Protein Length
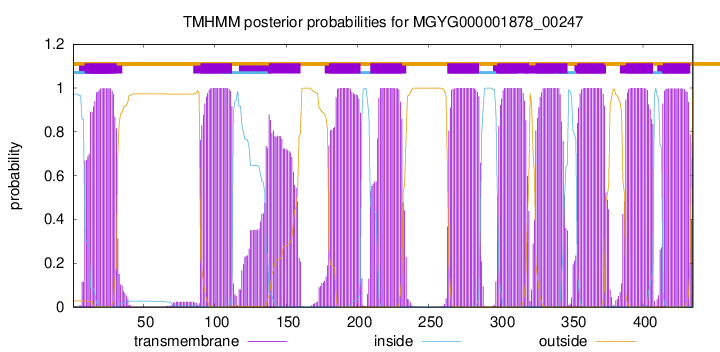
CGC
Molecular Weight
Isoelectric Point
435
49329.68
9.3419
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001878
3414068
MAG
Denmark
Europe
Gene Location
Start: 65663;
End: 66970
Strand: +
No EC number prediction in MGYG000001878_00247.
Family
Start
End
Evalue
family coverage
GT83
15
429
4e-43
0.7796296296296297
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG1807
ArnT
2.21e-17
18
422
13
418
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5EZM_A
9.76e-07
44
277
63
299
CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O67270
6.00e-10
30
340
20
320
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1
more
A8FRR0
5.52e-07
19
339
14
330
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Shewanella sediminis (strain HAW-EB3) OX=425104 GN=arnT PE=3 SV=1
more
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.973299
0.020810
0.001170
0.000050
0.000036
0.004661
start
end
9
31
90
112
138
160
180
202
209
231
263
285
298
320
325
347
354
373
388
407
414
433