You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001884_01351
You are here: Home > Sequence: MGYG000001884_01351
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1829 sp900760615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp900760615 | |||||||||||
| CAZyme ID | MGYG000001884_01351 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 590; End: 2419 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 71 | 215 | 1.2e-22 | 0.419953596287703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02449 | Glyco_hydro_42 | 2.31e-07 | 37 | 136 | 5 | 137 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| pfam01229 | Glyco_hydro_39 | 2.15e-06 | 108 | 247 | 122 | 285 | Glycosyl hydrolases family 39. |
| pfam11790 | Glyco_hydro_cc | 0.006 | 134 | 240 | 70 | 170 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| cd06152 | YjgF_YER057c_UK114_like_4 | 0.008 | 185 | 246 | 43 | 107 | YjgF, YER057c, and UK114 belong to a large family of proteins present in bacteria, archaea, and eukaryotes with no definitive function. The conserved domain is similar in structure to chorismate mutase but there is no sequence similarity and no functional connection. Members of this family have been implicated in isoleucine (Yeo7, Ibm1, aldR) and purine (YjgF) biosynthesis, as well as threonine anaerobic degradation (tdcF) and mitochondrial DNA maintenance (Ibm1). This domain homotrimerizes forming a distinct intersubunit cavity that may serve as a small molecule binding site. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 2.16e-144 | 16 | 607 | 18 | 609 |
| QDU57372.1 | 1.79e-93 | 19 | 604 | 22 | 613 |
| AVM44901.1 | 1.54e-51 | 19 | 572 | 19 | 589 |
| AHF90941.1 | 9.94e-48 | 25 | 568 | 232 | 804 |
| SDS89296.1 | 3.76e-31 | 21 | 244 | 300 | 530 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 2.30e-13 | 28 | 241 | 107 | 343 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
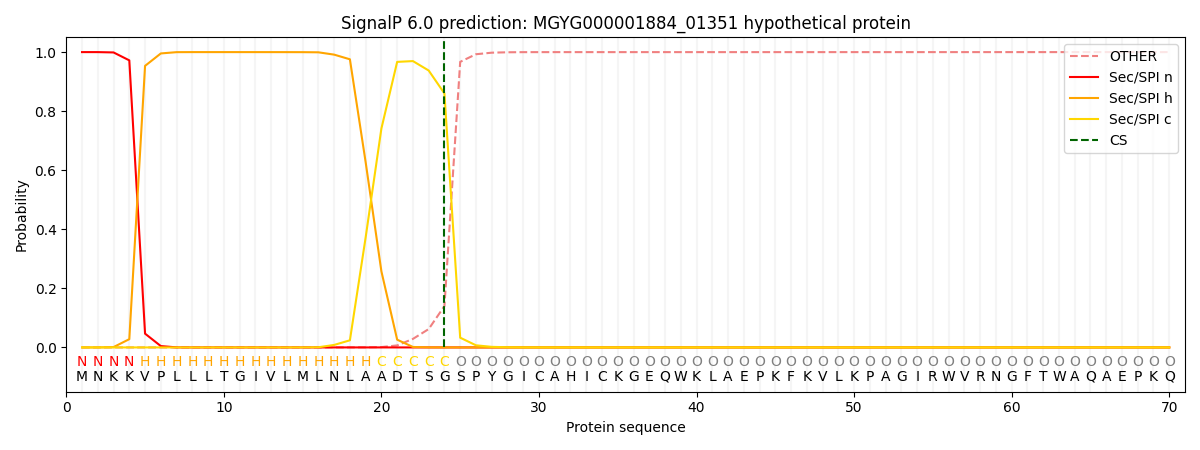
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000258 | 0.999103 | 0.000200 | 0.000139 | 0.000132 | 0.000135 |
