You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001885_00397
You are here: Home > Sequence: MGYG000001885_00397
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; | |||||||||||
| CAZyme ID | MGYG000001885_00397 | |||||||||||
| CAZy Family | GH38 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18144; End: 22097 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH38 | 27 | 291 | 2.9e-49 | 0.9219330855018587 |
| CBM32 | 1088 | 1204 | 5.1e-16 | 0.9193548387096774 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10789 | GH38N_AMII_ER_cytosolic | 7.08e-68 | 26 | 291 | 1 | 249 | N-terminal catalytic domain of endoplasmic reticulum(ER)/cytosolic class II alpha-mannosidases; glycoside hydrolase family 38 (GH38). The subfamily is represented by Saccharomyces cerevisiae vacuolar alpha-mannosidase Ams1, rat ER/cytosolic alpha-mannosidase Man2C1, and similar proteins. Members in this family share high sequence similarity. None of them have any classical signal sequence or membrane spanning domains, which are typical of sorting or targeting signals. Ams1 functions as a second resident vacuolar hydrolase in S. cerevisiae. It aids in recycling macromolecular components of the cell through hydrolysis of terminal, non-reducing alpha-d-mannose residues. Ams1 utilizes both the cytoplasm to vacuole targeting (Cvt, nutrient-rich conditions) and autophagic (starvation conditions) pathways for biosynthetic delivery to the vacuole. Man2C1is involved in oligosaccharide catabolism in both the ER and cytosol. It can catalyze the cobalt-dependent cleavage of alpha 1,2-, alpha 1,3-, and alpha 1,6-linked mannose residues. Members in this family are retaining glycosyl hydrolases of family GH38 that employs a two-step mechanism involving the formation of a covalent glycosyl-enzyme complex. Two carboxylic acids positioned within the active site act in concert: one as a catalytic nucleophile and the other as a general acid/base catalyst. |
| COG0383 | AMS1 | 6.30e-67 | 27 | 852 | 200 | 943 | Alpha-mannosidase [Carbohydrate transport and metabolism]. |
| pfam01074 | Glyco_hydro_38 | 3.31e-43 | 26 | 291 | 1 | 255 | Glycosyl hydrolases family 38 N-terminal domain. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. |
| cd10812 | GH38N_AMII_ScAms1_like | 8.40e-38 | 27 | 223 | 2 | 199 | N-terminal catalytic domain of yeast vacuolar alpha-mannosidases and similar proteins; glycoside hydrolase family 38 (GH38). The family is represented by Saccharomyces cerevisiae alpha-mannosidase (Ams1) and its eukaryotic homologs. Ams1 functions as a second resident vacuolar hydrolase in S. cerevisiae. It aids in recycling macromolecular components of the cell through hydrolysis of terminal, non-reducing alpha-d-mannose residues. Ams1 forms an oligomer in the cytoplasm and retains its oligomeric form during the import process. It utilizes both the Cvt (nutrient-rich conditions) and autophagic (starvation conditions) pathways for biosynthetic delivery to the vacuole. Mutants in either pathway are defective in Ams1 import. Members in this family show high sequence similarity with rat ER/cytosolic alpha-mannosidase Man2C1. |
| pfam07748 | Glyco_hydro_38C | 2.01e-34 | 509 | 703 | 1 | 198 | Glycosyl hydrolases family 38 C-terminal domain. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74823.1 | 0.0 | 5 | 1316 | 4 | 1313 |
| QDO68213.1 | 9.03e-260 | 14 | 1206 | 48 | 1235 |
| QUU01202.1 | 3.82e-259 | 20 | 1206 | 24 | 1203 |
| QCQ31060.1 | 8.06e-259 | 7 | 1206 | 8 | 1204 |
| BAD47088.1 | 1.60e-258 | 7 | 1206 | 8 | 1204 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6LZ1_A | 3.42e-58 | 23 | 854 | 279 | 1079 | Structureof S.pombe alpha-mannosidase Ams1 [Schizosaccharomyces pombe 972h-],6LZ1_B Structure of S.pombe alpha-mannosidase Ams1 [Schizosaccharomyces pombe 972h-],6LZ1_C Structure of S.pombe alpha-mannosidase Ams1 [Schizosaccharomyces pombe 972h-],6LZ1_D Structure of S.pombe alpha-mannosidase Ams1 [Schizosaccharomyces pombe 972h-] |
| 7DD9_A | 1.51e-57 | 23 | 867 | 279 | 1092 | ChainA, Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein [synthetic construct],7DD9_C Chain C, Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein [synthetic construct],7DD9_E Chain E, Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein [synthetic construct],7DD9_G Chain G, Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein [synthetic construct] |
| 5JM0_A | 5.22e-38 | 30 | 757 | 307 | 984 | Structureof the S. cerevisiae alpha-mannosidase 1 [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q54K67 | 5.09e-59 | 30 | 821 | 260 | 1058 | Alpha-mannosidase G OS=Dictyostelium discoideum OX=44689 GN=manG PE=1 SV=1 |
| Q9UT61 | 4.42e-57 | 23 | 852 | 279 | 1077 | Alpha-mannosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ams1 PE=1 SV=1 |
| Q9NTJ4 | 4.77e-57 | 34 | 796 | 260 | 981 | Alpha-mannosidase 2C1 OS=Homo sapiens OX=9606 GN=MAN2C1 PE=1 SV=1 |
| Q91W89 | 5.80e-55 | 26 | 796 | 251 | 980 | Alpha-mannosidase 2C1 OS=Mus musculus OX=10090 GN=Man2c1 PE=1 SV=1 |
| P21139 | 3.01e-53 | 34 | 796 | 259 | 981 | Alpha-mannosidase 2C1 OS=Rattus norvegicus OX=10116 GN=Man2c1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
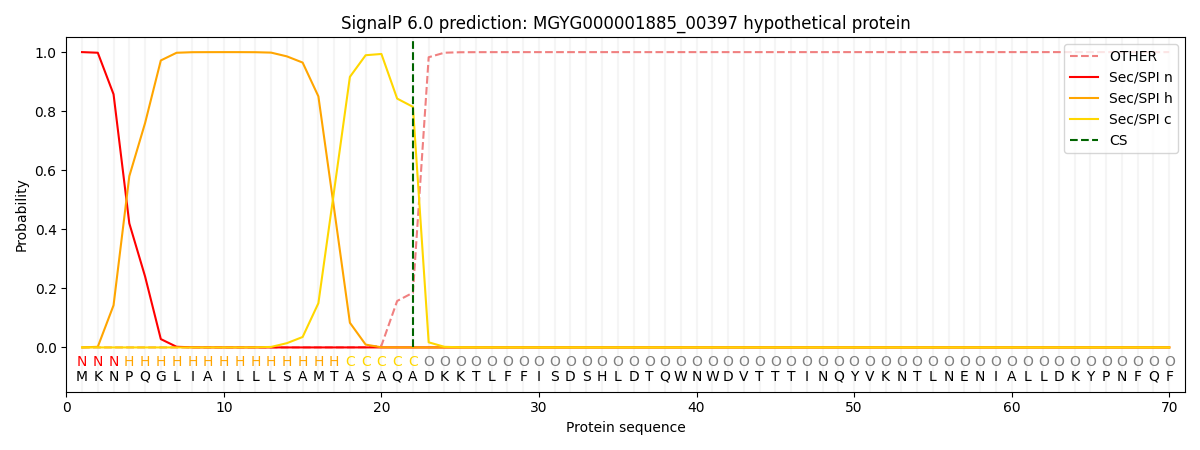
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000325 | 0.998927 | 0.000203 | 0.000192 | 0.000162 | 0.000149 |
