You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001886_02222
You are here: Home > Sequence: MGYG000001886_02222
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; | |||||||||||
| CAZyme ID | MGYG000001886_02222 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 169; End: 2718 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 169 | 610 | 6e-130 | 0.993421052631579 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 9.27e-28 | 164 | 455 | 1 | 313 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 4.02e-17 | 255 | 615 | 88 | 431 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02057 | Glyco_hydro_59 | 4.18e-05 | 201 | 480 | 25 | 251 | Glycosyl hydrolase family 59. |
| pfam14200 | RicinB_lectin_2 | 8.22e-05 | 658 | 739 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBE17217.1 | 1.60e-137 | 157 | 612 | 22 | 476 |
| AEW47939.1 | 4.18e-129 | 169 | 615 | 39 | 486 |
| AEW47948.1 | 4.18e-129 | 169 | 615 | 39 | 486 |
| ADF52950.1 | 7.15e-113 | 169 | 613 | 28 | 470 |
| AUX21860.1 | 1.20e-99 | 164 | 612 | 7 | 445 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CLW_A | 1.82e-17 | 167 | 616 | 8 | 500 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q76FP5 | 2.56e-46 | 162 | 552 | 20 | 409 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
| A0A401ETL2 | 4.58e-31 | 156 | 748 | 26 | 795 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
| Q4WBR2 | 1.71e-08 | 196 | 549 | 92 | 451 | Endo-1,6-beta-D-glucanase neg1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=neg1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
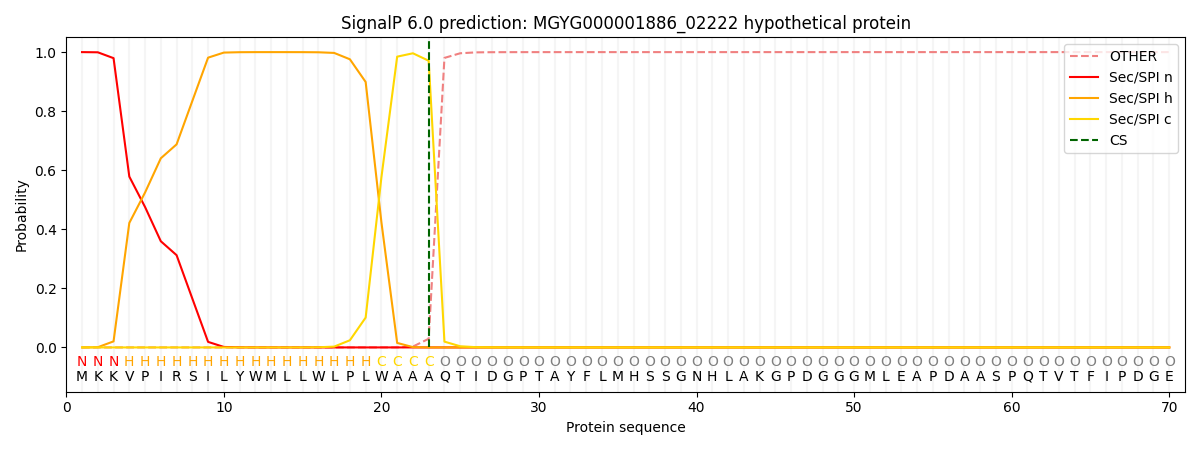
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000594 | 0.998478 | 0.000295 | 0.000202 | 0.000196 | 0.000202 |
