You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001887_00674
You are here: Home > Sequence: MGYG000001887_00674
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminiclostridium_E sp003512525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminiclostridium_E; Ruminiclostridium_E sp003512525 | |||||||||||
| CAZyme ID | MGYG000001887_00674 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 59016; End: 60440 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 166 | 441 | 9.9e-101 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 5.27e-54 | 166 | 442 | 16 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 7.53e-26 | 124 | 409 | 44 | 323 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL35063.1 | 8.86e-248 | 117 | 473 | 138 | 494 |
| CBK96026.1 | 3.98e-247 | 111 | 473 | 83 | 449 |
| CCO05659.1 | 9.17e-164 | 109 | 471 | 37 | 394 |
| CBL17363.1 | 3.90e-94 | 117 | 467 | 29 | 377 |
| BCN30798.1 | 1.18e-87 | 118 | 462 | 79 | 428 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6WQP_A | 4.34e-66 | 116 | 462 | 8 | 354 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 6XRK_A | 8.18e-65 | 123 | 466 | 23 | 373 | GH5-4broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate],6XRK_B GH5-4 broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate] |
| 6XSU_A | 9.82e-65 | 121 | 453 | 10 | 340 | GH5-4broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens],6XSU_B GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens] |
| 4X0V_A | 4.37e-63 | 126 | 462 | 42 | 393 | Structureof a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_B Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_C Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_D Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_E Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_F Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_G Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_H Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32] |
| 5D9N_A | 1.05e-62 | 123 | 463 | 12 | 350 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9N_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9P_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii],5D9P_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P28621 | 4.61e-58 | 111 | 466 | 30 | 376 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| O08342 | 2.56e-57 | 119 | 441 | 35 | 369 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P23660 | 2.96e-56 | 117 | 460 | 20 | 359 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| Q12647 | 5.43e-56 | 119 | 460 | 20 | 359 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P28623 | 3.56e-55 | 119 | 466 | 38 | 374 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
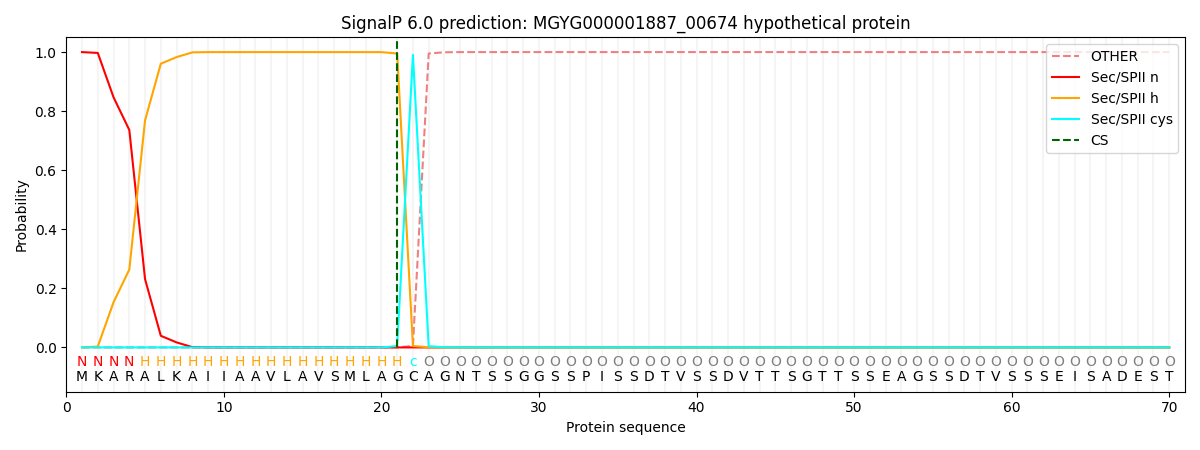
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000052 | 0.000000 | 0.000000 | 0.000000 |
