You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001887_00704
You are here: Home > Sequence: MGYG000001887_00704
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
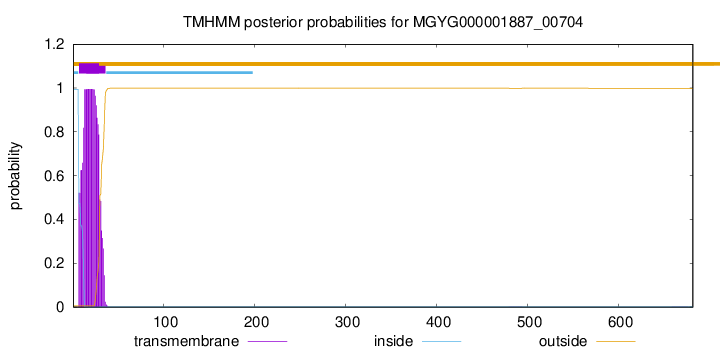
TMHMM annotations
Basic Information help
| Species | Ruminiclostridium_E sp003512525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminiclostridium_E; Ruminiclostridium_E sp003512525 | |||||||||||
| CAZyme ID | MGYG000001887_00704 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 96970; End: 99018 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 69 | 353 | 2.2e-102 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.11e-74 | 58 | 356 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.43e-31 | 28 | 381 | 30 | 387 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd00063 | FN3 | 3.65e-07 | 417 | 494 | 15 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| cd00063 | FN3 | 1.36e-06 | 496 | 587 | 1 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 2.13e-05 | 417 | 479 | 15 | 82 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL34359.1 | 1.35e-291 | 1 | 680 | 1 | 766 |
| CBK95964.1 | 6.87e-145 | 237 | 681 | 1 | 436 |
| CCO03822.1 | 1.51e-137 | 32 | 680 | 36 | 734 |
| AOZ96726.1 | 1.35e-129 | 37 | 386 | 35 | 394 |
| CBK74879.1 | 3.32e-129 | 5 | 386 | 3 | 396 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4NF7_A | 1.93e-130 | 37 | 386 | 4 | 363 | Crystalstructure of the GH5 family catalytic domain of Endo-1,4-beta-glucanase Cel5C from Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316] |
| 6XSU_A | 9.66e-110 | 40 | 378 | 12 | 342 | GH5-4broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens],6XSU_B GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens] |
| 4X0V_A | 1.27e-109 | 51 | 388 | 50 | 396 | Structureof a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_B Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_C Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_D Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_E Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_F Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_G Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_H Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32] |
| 5H4R_A | 3.56e-109 | 51 | 388 | 50 | 396 | thecomplex of Glycoside Hydrolase 5 Lichenase from Caldicellulosiruptor sp. F32 E188Q mutant and cellotetraose [Caldicellulosiruptor sp. F32] |
| 1EDG_A | 4.87e-104 | 51 | 386 | 32 | 374 | SingleCrystal Structure Determination Of The Catalytic Domain Of Celcca Carried Out At 15 Degree C [Ruminiclostridium cellulolyticum H10] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20847 | 4.17e-127 | 37 | 386 | 35 | 394 | Endoglucanase 1 OS=Butyrivibrio fibrisolvens OX=831 GN=end1 PE=3 SV=1 |
| P17901 | 5.10e-102 | 51 | 386 | 57 | 399 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P23661 | 8.70e-92 | 36 | 388 | 66 | 401 | Endoglucanase B OS=Ruminococcus albus OX=1264 GN=celB PE=3 SV=1 |
| P54937 | 2.81e-90 | 39 | 385 | 37 | 376 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P16216 | 1.30e-89 | 35 | 389 | 63 | 400 | Endoglucanase 1 OS=Ruminococcus albus OX=1264 GN=Eg I PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
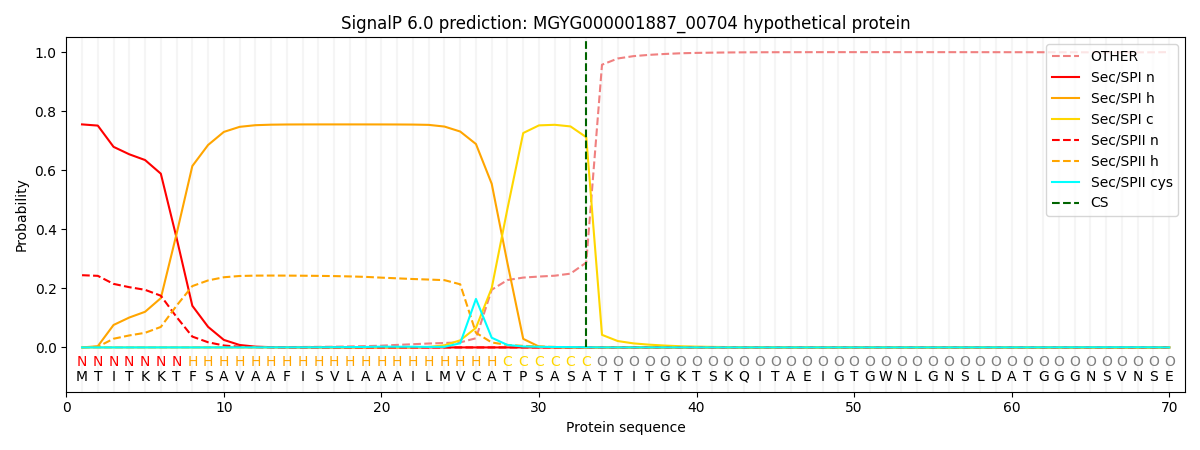
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000963 | 0.749234 | 0.249031 | 0.000294 | 0.000231 | 0.000206 |