You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001896_00263
Basic Information
help
| Species |
UMGS1840 sp900555525
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; UMGS1840; UMGS1840; UMGS1840; UMGS1840 sp900555525
|
| CAZyme ID |
MGYG000001896_00263
|
| CAZy Family |
GH141 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 713 |
|
81058.41 |
5.1508 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001896 |
2208881 |
MAG |
Denmark |
Europe |
|
| Gene Location |
Start: 22973;
End: 25114
Strand: +
|
No EC number prediction in MGYG000001896_00263.
| Family |
Start |
End |
Evalue |
family coverage |
| GH141 |
3 |
315 |
2.2e-27 |
0.538899430740038 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13229
|
Beta_helix |
4.34e-08 |
297 |
503 |
3 |
148 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229
|
Beta_helix |
9.33e-07 |
393 |
576 |
6 |
156 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
This protein is predicted as OTHER
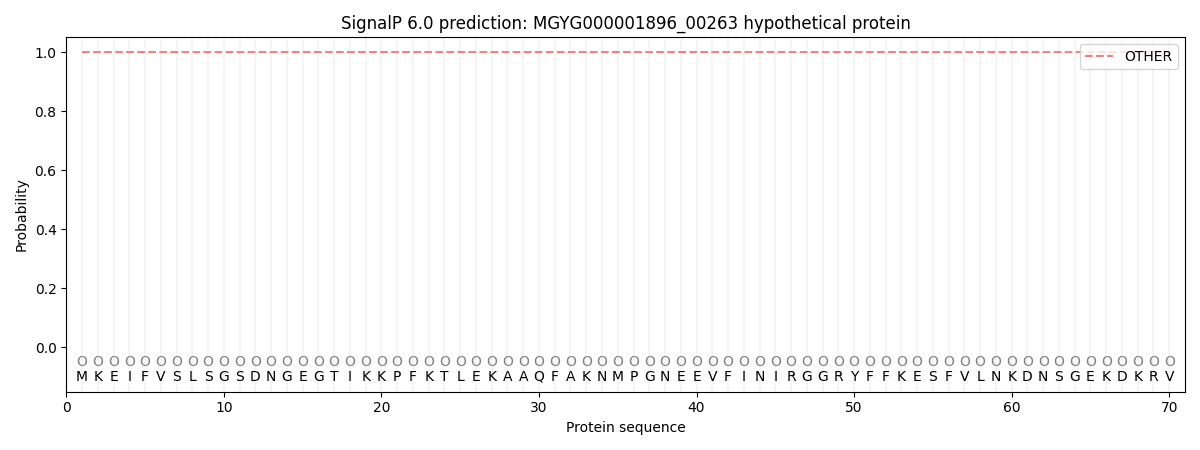
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000065
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000001896_00263.

