You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001903_00670
You are here: Home > Sequence: MGYG000001903_00670
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
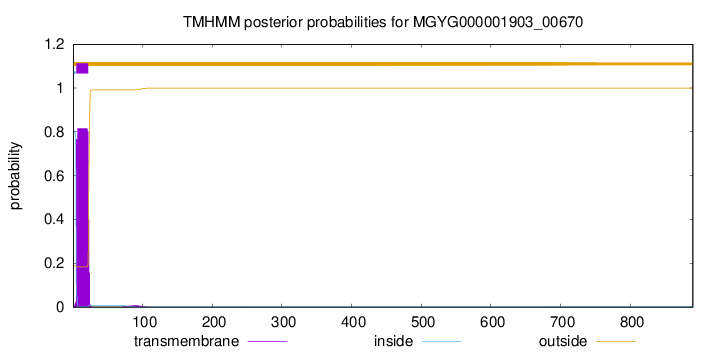
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; Firm-18; ; | |||||||||||
| CAZyme ID | MGYG000001903_00670 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7451; End: 10123 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 558 | 886 | 1.4e-41 | 0.9415384615384615 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 8.37e-12 | 558 | 880 | 118 | 466 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 1.58e-11 | 628 | 871 | 62 | 307 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| pfam07532 | Big_4 | 1.09e-06 | 180 | 233 | 5 | 59 | Bacterial Ig-like domain (group 4). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| PLN03010 | PLN03010 | 1.68e-06 | 627 | 871 | 141 | 381 | polygalacturonase |
| NF033190 | inl_like_NEAT_1 | 6.44e-05 | 247 | 375 | 587 | 705 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTH43708.1 | 1.44e-127 | 424 | 890 | 8 | 467 |
| AWW32777.1 | 2.49e-62 | 423 | 889 | 30 | 490 |
| SDS06655.1 | 1.87e-59 | 417 | 890 | 36 | 510 |
| ALJ04708.1 | 3.69e-59 | 419 | 836 | 41 | 438 |
| QIU92674.1 | 2.41e-58 | 423 | 873 | 29 | 467 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLP_A | 1.88e-10 | 642 | 799 | 195 | 354 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
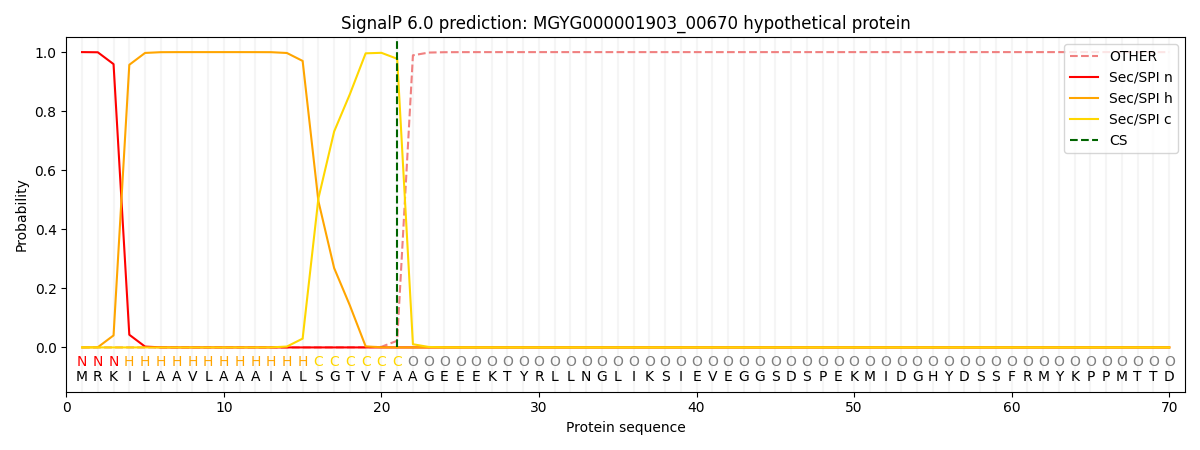
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000259 | 0.999043 | 0.000182 | 0.000165 | 0.000176 | 0.000152 |