You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001910_00156
You are here: Home > Sequence: MGYG000001910_00156
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella dubosii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella dubosii | |||||||||||
| CAZyme ID | MGYG000001910_00156 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 178021; End: 181902 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 351 | 650 | 1e-69 | 0.9826388888888888 |
| PL1 | 798 | 1001 | 5.6e-60 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02773 | PLN02773 | 3.65e-46 | 349 | 650 | 6 | 296 | pectinesterase |
| pfam01095 | Pectinesterase | 4.92e-43 | 352 | 650 | 4 | 295 | Pectinesterase. |
| PLN02432 | PLN02432 | 1.79e-40 | 338 | 650 | 1 | 286 | putative pectinesterase |
| PLN02990 | PLN02990 | 1.53e-36 | 352 | 622 | 263 | 527 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02301 | PLN02301 | 2.13e-35 | 352 | 619 | 240 | 500 | pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD41735.1 | 0.0 | 1 | 1293 | 1 | 1293 |
| QCP72287.1 | 0.0 | 1 | 1292 | 8 | 1316 |
| QCD38595.1 | 0.0 | 1 | 1292 | 8 | 1316 |
| ANU63102.1 | 0.0 | 29 | 1293 | 27 | 1294 |
| ASB38821.1 | 0.0 | 29 | 1293 | 27 | 1294 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XG2_A | 7.05e-28 | 347 | 612 | 2 | 258 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
| 3UW0_A | 1.15e-25 | 359 | 606 | 43 | 310 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 1GQ8_A | 1.25e-25 | 350 | 659 | 9 | 315 | Pectinmethylesterase from Carrot [Daucus carota] |
| 2NSP_A | 7.67e-19 | 360 | 658 | 18 | 342 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 1.01e-36 | 744 | 1203 | 21 | 414 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q5B297 | 1.91e-36 | 744 | 1205 | 21 | 413 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| Q2UB83 | 5.00e-35 | 744 | 1203 | 21 | 414 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q4WL88 | 1.69e-34 | 744 | 1203 | 22 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
| B0XMA2 | 5.60e-34 | 744 | 1203 | 22 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
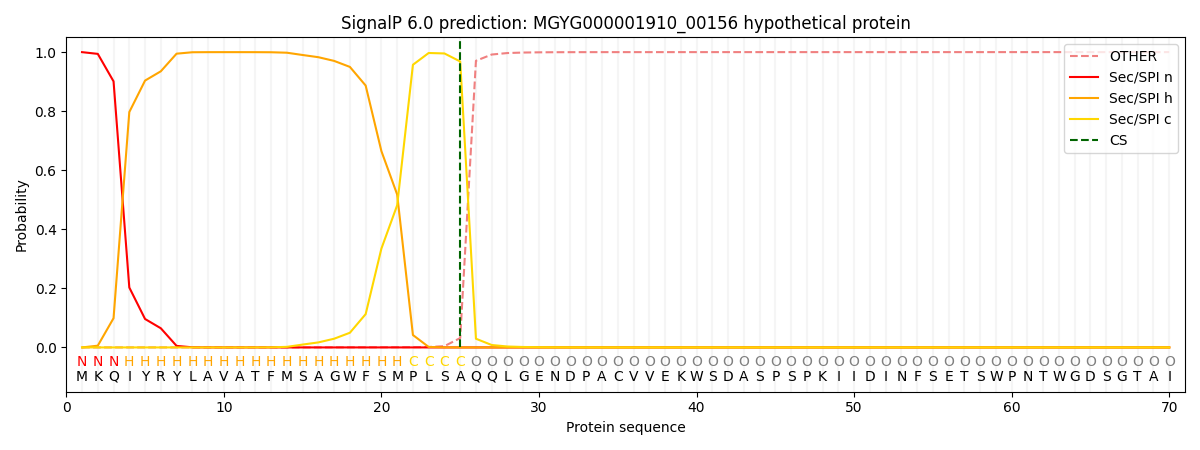
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000565 | 0.998520 | 0.000349 | 0.000206 | 0.000162 | 0.000147 |
