You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001910_00704
You are here: Home > Sequence: MGYG000001910_00704
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella dubosii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella dubosii | |||||||||||
| CAZyme ID | MGYG000001910_00704 | |||||||||||
| CAZy Family | GH138 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26556; End: 29249 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH138 | 9 | 897 | 0 | 0.9922822491730982 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02838 | Glyco_hydro_20b | 6.65e-05 | 23 | 106 | 21 | 100 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD40911.1 | 0.0 | 1 | 897 | 1 | 897 |
| AII68811.1 | 0.0 | 1 | 897 | 1 | 901 |
| QEW37250.1 | 0.0 | 1 | 897 | 1 | 901 |
| QUT83710.1 | 0.0 | 1 | 897 | 1 | 901 |
| AND20296.1 | 0.0 | 12 | 897 | 9 | 898 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6HZE_A | 0.0 | 12 | 897 | 10 | 893 | ChainA, BPa0997 [Phocaeicola paurosaccharolyticus],6HZE_B Chain B, BPa0997 [Phocaeicola paurosaccharolyticus],6HZG_A Chain A, BPa0997 N-ter E361S [Phocaeicola paurosaccharolyticus] |
| 6HZF_A | 0.0 | 12 | 897 | 10 | 893 | ChainA, BPa0997 [Phocaeicola paurosaccharolyticus] |
| 6HZF_B | 0.0 | 33 | 897 | 9 | 871 | ChainB, BPa0997 [Phocaeicola paurosaccharolyticus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
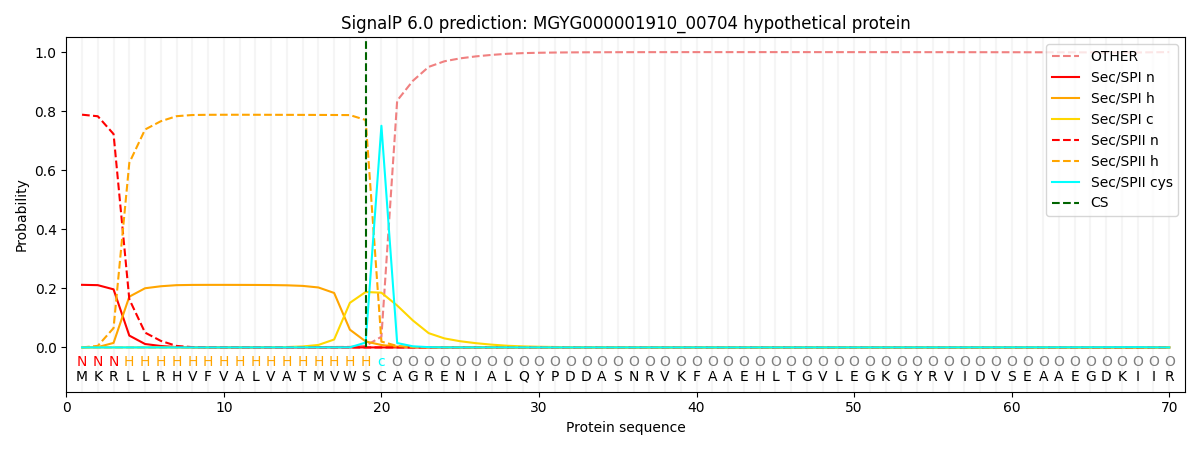
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000441 | 0.206889 | 0.792287 | 0.000115 | 0.000150 | 0.000123 |
