You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001918_00580
You are here: Home > Sequence: MGYG000001918_00580
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotellamassilia sp900541575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotellamassilia; Prevotellamassilia sp900541575 | |||||||||||
| CAZyme ID | MGYG000001918_00580 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37448; End: 41206 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 413 | 537 | 6.5e-16 | 0.8870967741935484 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 5.96e-15 | 414 | 537 | 7 | 126 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 3.04e-09 | 400 | 532 | 7 | 137 | Substituted updates: Jan 31, 2002 |
| cd14791 | GH36 | 5.86e-06 | 786 | 993 | 12 | 263 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFQ11998.1 | 0.0 | 30 | 1204 | 32 | 1017 |
| QFQ12302.1 | 2.21e-191 | 31 | 1199 | 805 | 1783 |
| SEH73474.1 | 1.24e-167 | 601 | 1199 | 247 | 822 |
| QWP25743.1 | 2.03e-162 | 550 | 1199 | 221 | 841 |
| QWP03233.1 | 2.03e-162 | 550 | 1199 | 221 | 841 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XLT_A | 2.43e-10 | 402 | 546 | 12 | 152 | ChainA, Galactose oxidase [Fusarium graminearum] |
| 6XLR_A | 2.43e-10 | 402 | 546 | 12 | 152 | ChainA, Galactose oxidase [Fusarium graminearum],6XLS_A Chain A, Galactose oxidase [Fusarium graminearum] |
| 2WQ8_A | 2.47e-10 | 402 | 546 | 33 | 173 | ChainA, GALACTOSE OXIDASE [Fusarium graminearum] |
| 2EIC_A | 4.17e-10 | 402 | 546 | 11 | 151 | ChainA, Galactose oxidase [Fusarium graminearum] |
| 2EIB_A | 4.17e-10 | 402 | 546 | 11 | 151 | ChainA, Galactose oxidase [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| I1S2N3 | 1.81e-09 | 402 | 546 | 52 | 192 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
| P0CS93 | 2.38e-09 | 402 | 546 | 52 | 192 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
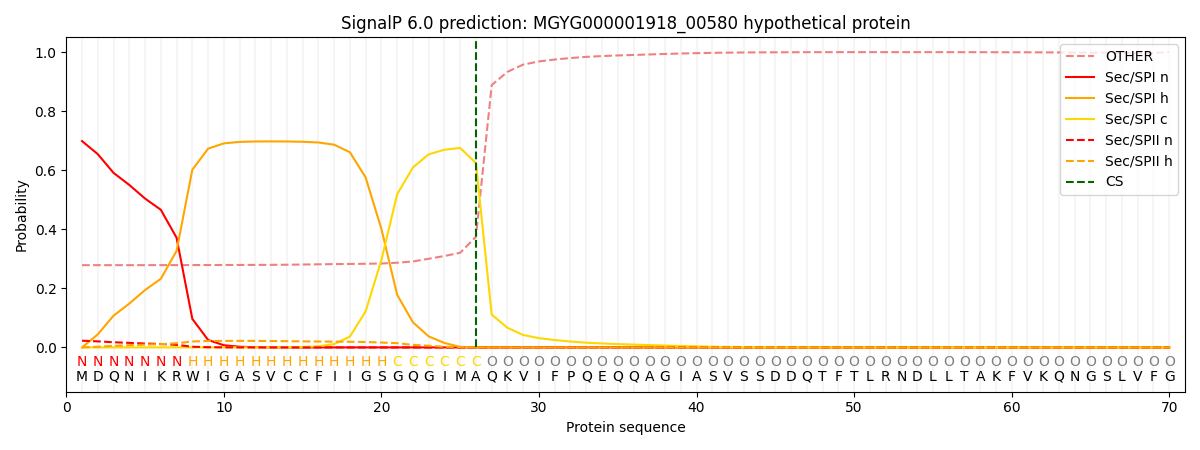
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.299182 | 0.674671 | 0.024431 | 0.000695 | 0.000399 | 0.000594 |
