You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001921_01756
You are here: Home > Sequence: MGYG000001921_01756
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; | |||||||||||
| CAZyme ID | MGYG000001921_01756 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 83500; End: 85851 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 19 | 529 | 7.1e-61 | 0.9867172675521821 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13180 | PDZ_2 | 3.83e-09 | 714 | 781 | 2 | 72 | PDZ domain. |
| cd00987 | PDZ_serine_protease | 4.04e-08 | 712 | 781 | 18 | 90 | PDZ domain of trypsin-like serine proteases, such as DegP/HtrA, which are oligomeric proteins involved in heat-shock response, chaperone function, and apoptosis. May be responsible for substrate recognition and/or binding, as most PDZ domains bind C-terminal polypeptides, though binding to internal (non-C-terminal) polypeptides and even to lipids has been demonstrated. In this subfamily of protease-associated PDZ domains a C-terminal beta-strand forms the peptide-binding groove base, a circular permutation with respect to PDZ domains found in Eumetazoan signaling proteins. |
| cd00989 | PDZ_metalloprotease | 5.59e-08 | 707 | 783 | 1 | 79 | PDZ domain of bacterial and plant zinc metalloprotases, presumably membrane-associated or integral membrane proteases, which may be involved in signalling and regulatory mechanisms. May be responsible for substrate recognition and/or binding, as most PDZ domains bind C-terminal polypeptides, and binding to internal (non-C-terminal) polypeptides and even to lipids has been demonstrated. In this subfamily of protease-associated PDZ domains a C-terminal beta-strand forms the peptide-binding groove base, a circular permutation with respect to PDZ domains found in Eumetazoan signaling proteins. |
| smart00228 | PDZ | 5.07e-07 | 696 | 773 | 4 | 84 | Domain present in PSD-95, Dlg, and ZO-1/2. Also called DHR (Dlg homologous region) or GLGF (relatively well conserved tetrapeptide in these domains). Some PDZs have been shown to bind C-terminal polypeptides; others appear to bind internal (non-C-terminal) polypeptides. Different PDZs possess different binding specificities. |
| pfam13229 | Beta_helix | 1.03e-06 | 305 | 457 | 1 | 136 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWW32784.1 | 2.47e-192 | 16 | 762 | 29 | 775 |
| QTD80365.1 | 7.45e-107 | 1 | 159 | 1 | 159 |
| QWO88128.1 | 7.45e-107 | 1 | 159 | 1 | 159 |
| QUF77596.1 | 7.45e-107 | 1 | 159 | 1 | 159 |
| QEE55114.1 | 7.45e-107 | 1 | 159 | 1 | 159 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 5.73e-18 | 19 | 528 | 26 | 580 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
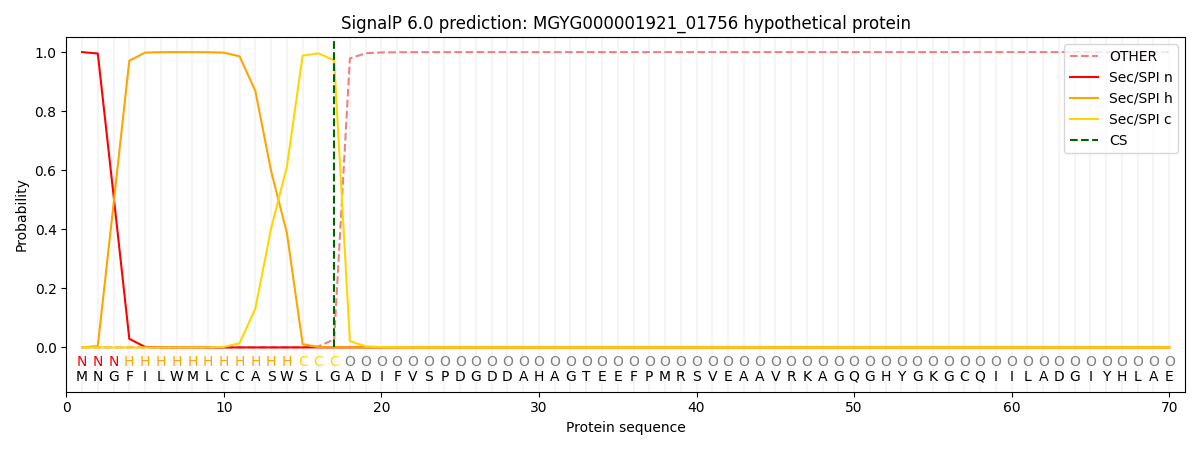
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000450 | 0.998726 | 0.000271 | 0.000180 | 0.000181 | 0.000176 |
