You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001925_00063
You are here: Home > Sequence: MGYG000001925_00063
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900544675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900544675 | |||||||||||
| CAZyme ID | MGYG000001925_00063 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 66230; End: 68476 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 504 | 715 | 3.2e-16 | 0.8678414096916299 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00930 | DPPIV_N | 3.84e-49 | 120 | 455 | 18 | 352 | Dipeptidyl peptidase IV (DPP IV) N-terminal region. This family is an alignment of the region to the N-terminal side of the active site. The Prosite motif does not correspond to this Pfam entry. |
| pfam00326 | Peptidase_S9 | 2.44e-43 | 547 | 733 | 11 | 210 | Prolyl oligopeptidase family. |
| COG1506 | DAP2 | 7.13e-37 | 156 | 714 | 58 | 594 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG0412 | DLH | 4.14e-10 | 546 | 744 | 50 | 226 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG1073 | FrsA | 6.01e-06 | 588 | 734 | 147 | 299 | Fermentation-respiration switch protein FrsA, has esterase activity, DUF1100 family [Signal transduction mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AJY87077.1 | 1.06e-196 | 69 | 742 | 270 | 950 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2D5L_A | 1.90e-52 | 157 | 716 | 120 | 686 | CrystalStructure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis [Porphyromonas gingivalis W83],2EEP_A Prolyl Tripeptidyl Aminopeptidase Complexed with an Inhibitor [Porphyromonas gingivalis W83] |
| 2Z3W_A | 3.52e-52 | 157 | 716 | 120 | 686 | ChainA, Dipeptidyl aminopeptidase IV [Porphyromonas gingivalis W83],2Z3Z_A Chain A, Dipeptidyl aminopeptidase IV [Porphyromonas gingivalis W83] |
| 2DCM_A | 4.79e-52 | 157 | 716 | 120 | 686 | ChainA, dipeptidyl aminopeptidase IV, putative [Porphyromonas gingivalis W83] |
| 2ECF_A | 2.45e-51 | 166 | 724 | 160 | 727 | CrystalStructure of Dipeptidyl Aminopeptidase IV from Stenotrophomonas maltophilia [Stenotrophomonas maltophilia] |
| 5YP1_A | 6.41e-51 | 119 | 733 | 125 | 743 | Crystalstructure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP1_B Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP1_C Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP1_D Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP2_A Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP2_B Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP3_A Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana [Pseudoxanthomonas mexicana],5YP3_B Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana [Pseudoxanthomonas mexicana],5YP3_C Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana [Pseudoxanthomonas mexicana],5YP3_D Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana [Pseudoxanthomonas mexicana],5YP4_A Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP4_B Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP4_C Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP4_D Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7MUW6 | 1.42e-51 | 157 | 716 | 146 | 712 | Prolyl tripeptidyl peptidase OS=Porphyromonas gingivalis (strain ATCC BAA-308 / W83) OX=242619 GN=ptpA PE=1 SV=1 |
| B2RJX3 | 4.85e-51 | 157 | 716 | 146 | 712 | Prolyl tripeptidyl peptidase OS=Porphyromonas gingivalis (strain ATCC 33277 / DSM 20709 / CIP 103683 / JCM 12257 / NCTC 11834 / 2561) OX=431947 GN=ptpA PE=3 SV=1 |
| Q6F3I7 | 3.51e-50 | 119 | 733 | 125 | 743 | Dipeptidyl aminopeptidase 4 OS=Pseudoxanthomonas mexicana OX=128785 GN=dap4 PE=1 SV=1 |
| A2QEK7 | 2.34e-31 | 52 | 746 | 153 | 881 | Probable dipeptidyl-aminopeptidase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=dapB PE=3 SV=1 |
| B0XYK8 | 2.41e-31 | 112 | 716 | 239 | 869 | Probable dipeptidyl-aminopeptidase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=dapB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
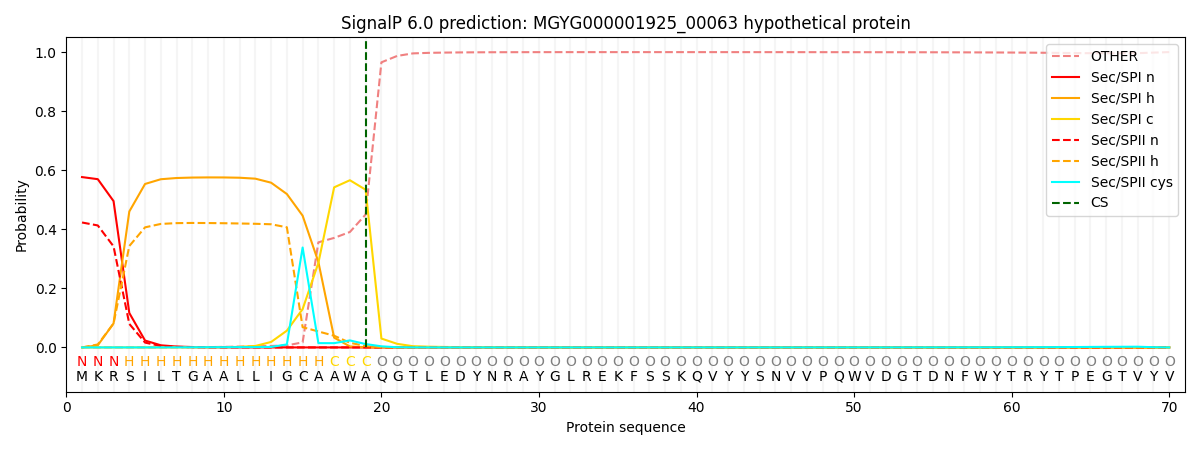
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000947 | 0.563951 | 0.434147 | 0.000408 | 0.000295 | 0.000231 |
