You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001925_02163
You are here: Home > Sequence: MGYG000001925_02163
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900544675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900544675 | |||||||||||
| CAZyme ID | MGYG000001925_02163 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27434; End: 30232 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 19 | 703 | 3.1e-58 | 0.7220744680851063 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.84e-22 | 21 | 428 | 9 | 431 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00703 | Glyco_hydro_2 | 2.76e-11 | 176 | 282 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10150 | PRK10150 | 2.71e-08 | 68 | 405 | 63 | 417 | beta-D-glucuronidase; Provisional |
| pfam02836 | Glyco_hydro_2_C | 3.15e-07 | 284 | 405 | 1 | 135 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK09525 | lacZ | 4.74e-07 | 237 | 417 | 288 | 476 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY36269.1 | 0.0 | 1 | 932 | 1 | 932 |
| QUT92878.1 | 0.0 | 22 | 929 | 28 | 936 |
| ALJ61552.1 | 0.0 | 22 | 929 | 28 | 936 |
| QUT34134.1 | 0.0 | 22 | 929 | 26 | 936 |
| QBJ17536.1 | 0.0 | 22 | 929 | 26 | 936 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6NCW_A | 1.44e-16 | 67 | 419 | 51 | 392 | Crystalstructure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_B Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_C Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_D Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCX_A Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_B Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_C Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_D Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi] |
| 6XXW_A | 4.29e-15 | 19 | 424 | 27 | 439 | Structureof beta-D-Glucuronidase for Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 7VQM_A | 5.59e-09 | 60 | 422 | 84 | 425 | ChainA, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_B Chain B, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_C Chain C, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_D Chain D, GH2 beta-galacturonate AqGalA [Aquimarina sp.] |
| 5C70_A | 9.76e-09 | 63 | 418 | 64 | 435 | Thestructure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae],5C70_B The structure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae] |
| 6EC6_A | 9.90e-09 | 68 | 419 | 90 | 455 | Ruminococcusgnavus Beta-glucuronidase [[Ruminococcus] gnavus],6EC6_B Ruminococcus gnavus Beta-glucuronidase [[Ruminococcus] gnavus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q47077 | 2.00e-07 | 99 | 417 | 139 | 477 | Beta-galactosidase OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
| Q2XQU3 | 4.53e-07 | 99 | 417 | 139 | 477 | Beta-galactosidase 2 OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
| A1JTC4 | 1.03e-06 | 52 | 417 | 116 | 491 | Beta-galactosidase OS=Yersinia enterocolitica serotype O:8 / biotype 1B (strain NCTC 13174 / 8081) OX=393305 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
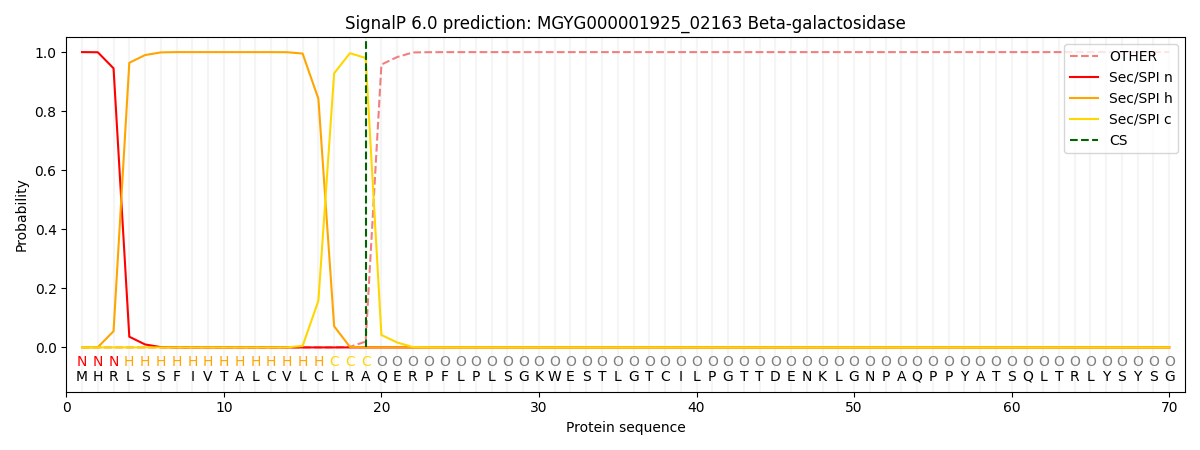
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000182 | 0.999246 | 0.000149 | 0.000142 | 0.000131 | 0.000128 |
