You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001927_01161
You are here: Home > Sequence: MGYG000001927_01161
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
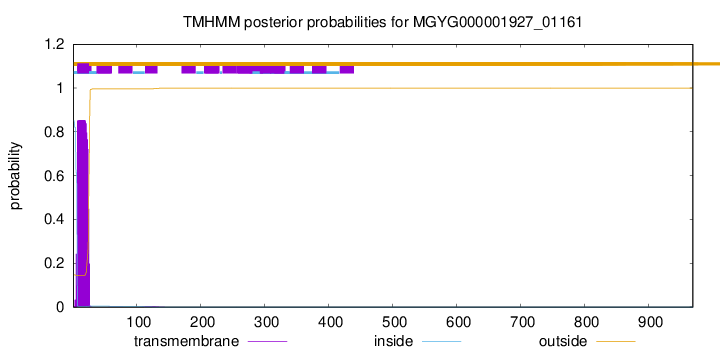
TMHMM annotations
Basic Information help
| Species | Prevotella sp900547005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900547005 | |||||||||||
| CAZyme ID | MGYG000001927_01161 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34959; End: 37868 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 480 | 644 | 6.9e-34 | 0.8168316831683168 |
| CBM77 | 865 | 959 | 2.1e-17 | 0.9029126213592233 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 4.96e-35 | 341 | 777 | 1 | 345 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam18283 | CBM77 | 1.05e-22 | 864 | 966 | 6 | 108 | Carbohydrate binding module 77. This domain is the non-catalytic carbohydrate binding module 77 (CBM77) present in Ruminococcus flavefaciens. CBMs fulfil a critical targeting function in plant cell wall depolymerisation. In CBM77, a cluster of conserved basic residues (Lys1092, Lys1107 and Lys1162) confer calcium-independent recognition of homogalacturonan. |
| smart00656 | Amb_all | 2.85e-21 | 483 | 646 | 17 | 190 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 1.06e-07 | 481 | 642 | 33 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| pfam13229 | Beta_helix | 0.005 | 552 | 685 | 22 | 147 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOR20273.1 | 8.31e-183 | 269 | 807 | 35 | 551 |
| CBW15186.1 | 1.38e-182 | 269 | 807 | 50 | 566 |
| QOR18408.1 | 1.24e-181 | 268 | 807 | 63 | 580 |
| QCD40816.1 | 2.40e-180 | 225 | 815 | 10 | 588 |
| QCP73706.1 | 2.40e-180 | 225 | 815 | 10 | 588 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1PCL_A | 1.77e-09 | 432 | 642 | 33 | 276 | ChainA, PECTATE LYASE E [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1CYB8 | 3.05e-12 | 483 | 697 | 93 | 298 | Probable pectate lyase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyA PE=3 SV=1 |
| B0XT32 | 5.48e-12 | 483 | 697 | 93 | 298 | Probable pectate lyase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyA PE=3 SV=1 |
| Q4WIT0 | 5.48e-12 | 483 | 697 | 93 | 298 | Probable pectate lyase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyA PE=3 SV=1 |
| A2QV36 | 1.83e-10 | 483 | 699 | 95 | 302 | Probable pectate lyase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=plyA PE=3 SV=1 |
| Q9C2Z0 | 1.83e-10 | 483 | 699 | 95 | 302 | Pectate lyase A OS=Aspergillus niger OX=5061 GN=plyA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
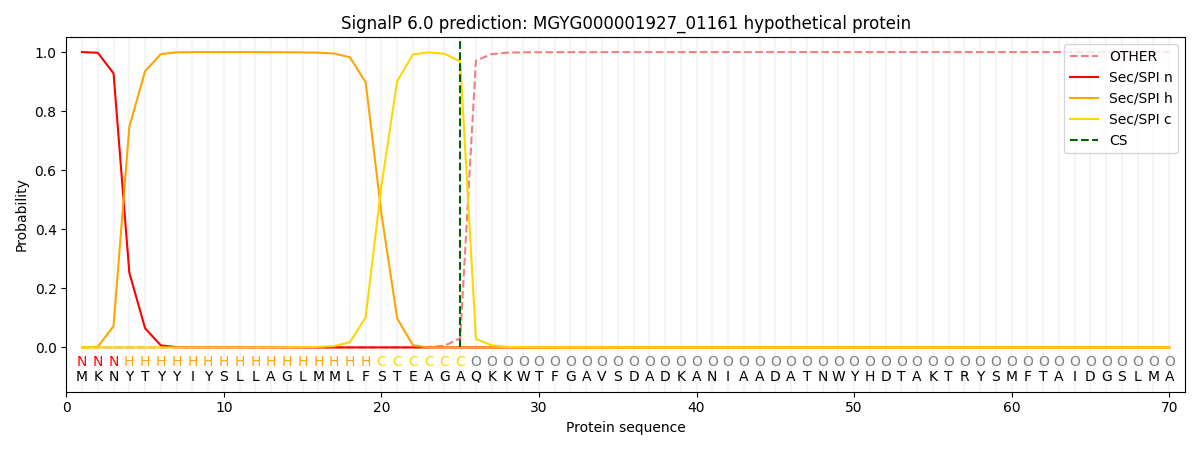
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000495 | 0.998681 | 0.000214 | 0.000214 | 0.000195 | 0.000175 |