You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001929_00123
You are here: Home > Sequence: MGYG000001929_00123
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paramuribaculum sp900752995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Paramuribaculum; Paramuribaculum sp900752995 | |||||||||||
| CAZyme ID | MGYG000001929_00123 | |||||||||||
| CAZy Family | GH29 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 142503; End: 146174 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH29 | 23 | 374 | 4.9e-93 | 0.953757225433526 |
| GH3 | 569 | 793 | 9.9e-68 | 0.9814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 3.93e-178 | 457 | 1221 | 11 | 763 | beta-glucosidase BglX. |
| pfam01120 | Alpha_L_fucos | 7.12e-108 | 35 | 371 | 3 | 333 | Alpha-L-fucosidase. |
| smart00812 | Alpha_L_fucos | 7.14e-94 | 34 | 405 | 3 | 372 | Alpha-L-fucosidase. O-Glycosyl hydrolases (EC 3.2.1.-) are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycosyl hydrolases, based on sequence similarity, has led to the definition of 85 different families. This classification is available on the CAZy (CArbohydrate-Active EnZymes) web site. Because the fold of proteins is better conserved than their sequences, some of the families can be grouped in 'clans'. Family 29 encompasses alpha-L-fucosidases, which is a lysosomal enzyme responsible for hydrolyzing the alpha-1,6-linked fucose joined to the reducing-end N-acetylglucosamine of the carbohydrate moieties of glycoproteins. Deficiency of alpha-L-fucosidase results in the lysosomal storage disease fucosidosis. |
| COG1472 | BglX | 5.34e-87 | 488 | 933 | 1 | 397 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PLN03080 | PLN03080 | 2.72e-83 | 572 | 1208 | 78 | 769 | Probable beta-xylosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADV43485.1 | 2.17e-304 | 468 | 1222 | 24 | 781 |
| QEW35133.1 | 4.73e-299 | 469 | 1220 | 29 | 783 |
| ABR38266.1 | 6.70e-299 | 469 | 1220 | 29 | 783 |
| QQY39447.1 | 7.19e-299 | 469 | 1220 | 31 | 785 |
| SCD22149.1 | 1.13e-285 | 470 | 1223 | 26 | 782 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z87_A | 8.68e-140 | 470 | 1215 | 36 | 778 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 5YOT_A | 4.33e-134 | 468 | 1223 | 5 | 757 | Isoprimeverose-producingenzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YOT_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_A Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40] |
| 7EAP_A | 3.20e-133 | 468 | 1223 | 5 | 757 | ChainA, Fn3_like domain-containing protein [Aspergillus oryzae RIB40] |
| 5XXL_A | 4.19e-130 | 477 | 1215 | 10 | 745 | Crystalstructure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXL_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXM_A Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482],5XXM_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482] |
| 5XXN_A | 2.20e-129 | 477 | 1215 | 10 | 745 | CrystalStructure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose [Bacteroides thetaiotaomicron VPI-5482],5XXN_B Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose [Bacteroides thetaiotaomicron VPI-5482],5XXO_A Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose [Bacteroides thetaiotaomicron VPI-5482],5XXO_B Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56078 | 1.84e-130 | 457 | 1222 | 11 | 764 | Periplasmic beta-glucosidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bglX PE=3 SV=2 |
| P33363 | 3.58e-130 | 457 | 1222 | 11 | 764 | Periplasmic beta-glucosidase OS=Escherichia coli (strain K12) OX=83333 GN=bglX PE=3 SV=2 |
| T2KMH0 | 3.10e-129 | 572 | 1217 | 62 | 716 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
| T2KMH9 | 3.22e-122 | 470 | 1223 | 29 | 755 | Putative beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22230 PE=2 SV=1 |
| A7LXU3 | 1.36e-87 | 477 | 1213 | 35 | 767 | Beta-glucosidase BoGH3B OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02659 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
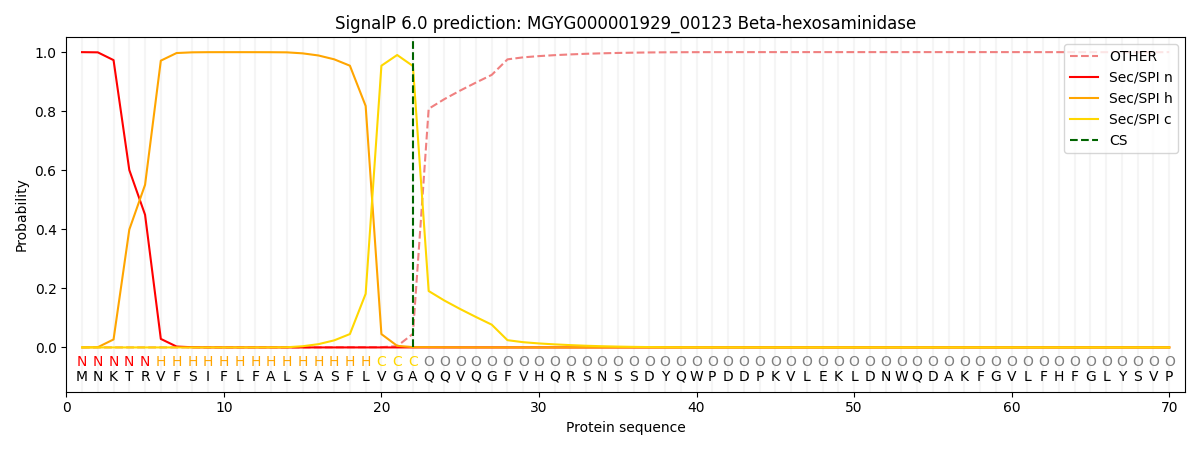
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001288 | 0.997349 | 0.000475 | 0.000295 | 0.000283 | 0.000268 |

