You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001929_01670
You are here: Home > Sequence: MGYG000001929_01670
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paramuribaculum sp900752995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Paramuribaculum; Paramuribaculum sp900752995 | |||||||||||
| CAZyme ID | MGYG000001929_01670 | |||||||||||
| CAZy Family | GH89 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12884; End: 15028 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH89 | 60 | 707 | 2.9e-224 | 0.9939668174962293 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05089 | NAGLU | 6.62e-165 | 117 | 434 | 1 | 333 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This central domain has a tim barrel fold. |
| pfam12972 | NAGLU_C | 6.34e-92 | 442 | 702 | 1 | 257 | Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This C-terminal domain has an all alpha helical fold. |
| pfam12971 | NAGLU_N | 1.61e-19 | 24 | 102 | 1 | 81 | Alpha-N-acetylglucosaminidase (NAGLU) N-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This N-terminal domain has an alpha-beta fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50280.1 | 6.12e-259 | 21 | 697 | 17 | 703 |
| QRO24616.1 | 6.97e-253 | 26 | 705 | 22 | 711 |
| AUI45172.1 | 4.51e-246 | 9 | 706 | 6 | 714 |
| QCQ35767.1 | 4.51e-246 | 9 | 706 | 6 | 714 |
| QKH85154.1 | 6.39e-246 | 9 | 706 | 6 | 714 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VC9_A | 2.18e-133 | 51 | 683 | 202 | 846 | Family89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin [Clostridium perfringens],2VCA_A Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine [Clostridium perfringens],2VCB_A Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc [Clostridium perfringens],2VCC_A Family 89 Glycoside Hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 7MFK_A | 2.66e-133 | 51 | 683 | 210 | 854 | ChainA, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124],7MFL_A Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124] |
| 4A4A_A | 2.93e-132 | 51 | 683 | 225 | 869 | CpGH89(E483Q, E601Q), from Clostridium perfringens, in complex with its substrate GlcNAc-alpha-1,4-galactose [Clostridium perfringens] |
| 4XWH_A | 1.00e-107 | 20 | 702 | 3 | 704 | Crystalstructure of the human N-acetyl-alpha-glucosaminidase [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54802 | 9.68e-107 | 20 | 702 | 26 | 727 | Alpha-N-acetylglucosaminidase OS=Homo sapiens OX=9606 GN=NAGLU PE=1 SV=2 |
| Q9FNA3 | 4.28e-98 | 14 | 685 | 37 | 768 | Alpha-N-acetylglucosaminidase OS=Arabidopsis thaliana OX=3702 GN=NAGLU PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
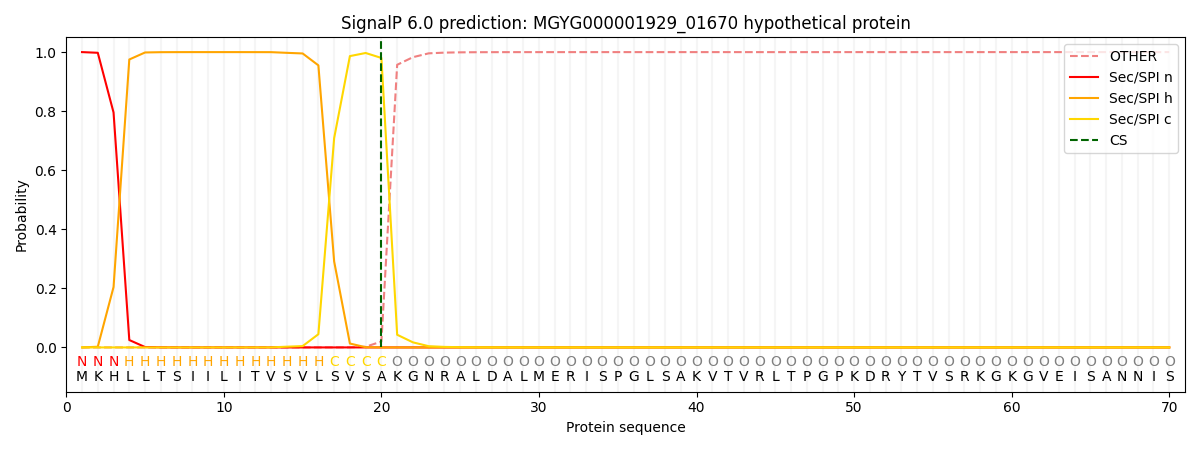
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000317 | 0.999002 | 0.000200 | 0.000149 | 0.000157 | 0.000150 |
