You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001948_00314
You are here: Home > Sequence: MGYG000001948_00314
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Muribaculum sp002492595 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum sp002492595 | |||||||||||
| CAZyme ID | MGYG000001948_00314 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 148926; End: 151160 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 483 | 739 | 3.9e-49 | 0.6798679867986799 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 6.83e-37 | 483 | 734 | 54 | 260 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 6.62e-35 | 487 | 734 | 100 | 305 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.46e-23 | 481 | 734 | 117 | 334 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 1.76e-10 | 60 | 147 | 5 | 92 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 7.30e-06 | 98 | 148 | 1 | 50 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD34661.1 | 0.0 | 1 | 744 | 1 | 766 |
| AXH21505.1 | 1.25e-221 | 3 | 742 | 2 | 744 |
| EDV05072.1 | 1.25e-221 | 3 | 742 | 2 | 744 |
| QRQ48288.1 | 3.57e-214 | 1 | 742 | 7 | 717 |
| QUT46089.1 | 3.26e-212 | 1 | 742 | 7 | 717 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGS_A | 2.29e-26 | 156 | 303 | 1 | 149 | BiXyn10ACBM1 APO [Bacteroides intestinalis DSM 17393] |
| 4QPW_A | 3.11e-23 | 165 | 301 | 4 | 141 | BiXyn10ACBM1 with Xylohexaose Bound [Bacteroides intestinalis DSM 17393] |
| 5Y3X_A | 2.83e-16 | 471 | 734 | 122 | 351 | Crystalstructure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_B Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_C Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_D Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_E Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_F Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL] |
| 1CLX_A | 1.98e-15 | 504 | 742 | 117 | 345 | CatalyticCore Of Xylanase A [Cellvibrio japonicus],1CLX_B Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_C Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_D Catalytic Core Of Xylanase A [Cellvibrio japonicus] |
| 1W2V_A | 2.00e-15 | 504 | 742 | 118 | 346 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W2V_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23556 | 2.26e-16 | 481 | 733 | 111 | 335 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=xynA PE=1 SV=1 |
| O94163 | 2.67e-15 | 485 | 741 | 126 | 327 | Endo-1,4-beta-xylanase F1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF1 PE=1 SV=1 |
| P14768 | 3.85e-14 | 504 | 742 | 381 | 609 | Endo-1,4-beta-xylanase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynA PE=1 SV=2 |
| Q4P902 | 6.06e-14 | 484 | 742 | 136 | 340 | Endo-1,4-beta-xylanase UM03411 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=UMAG_03411 PE=1 SV=1 |
| A1CHQ0 | 1.47e-13 | 485 | 741 | 126 | 317 | Probable endo-1,4-beta-xylanase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=xlnC PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
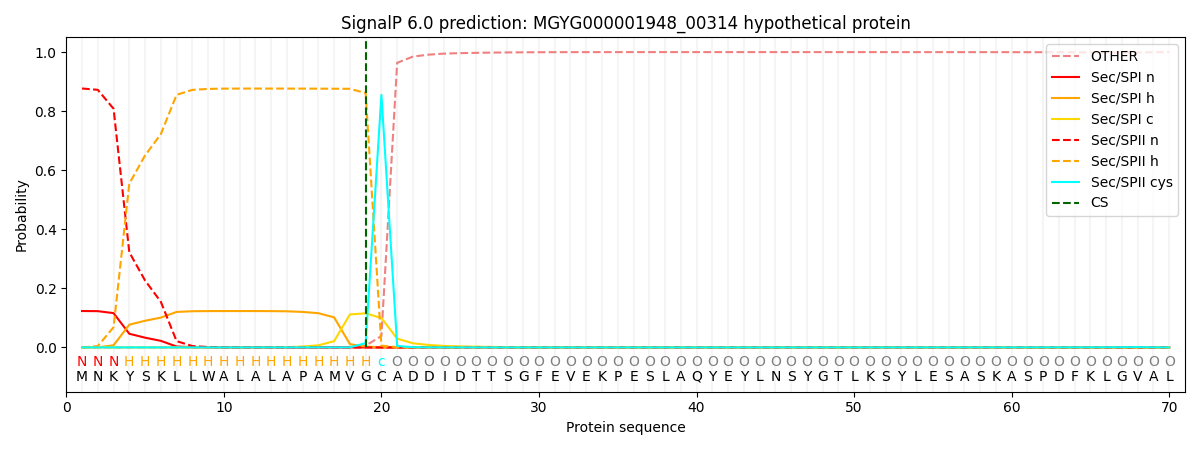
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000219 | 0.120307 | 0.879278 | 0.000065 | 0.000078 | 0.000069 |
