You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001951_01391
You are here: Home > Sequence: MGYG000001951_01391
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
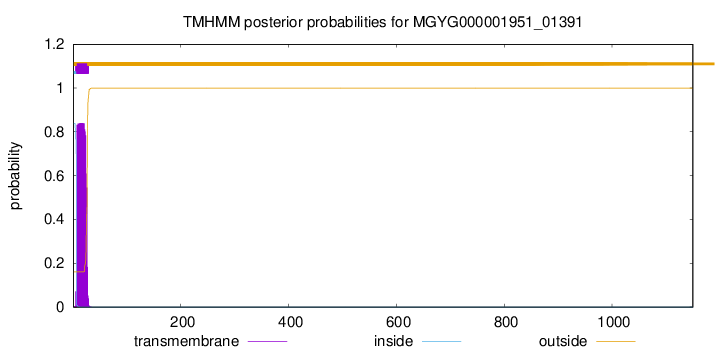
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900548975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900548975 | |||||||||||
| CAZyme ID | MGYG000001951_01391 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16652; End: 20104 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 210 | 737 | 2.3e-150 | 0.6759708737864077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 34 | 701 | 3 | 866 | alpha-L-rhamnosidase. |
| pfam11875 | DUF3395 | 7.54e-08 | 760 | 833 | 55 | 143 | Domain of unknown function (DUF3395). This domain is functionally uncharacterized. This domain is found in eukaryotes. This presumed domain is typically between 147 to 176 amino acids in length. This domain is found associated with pfam00226. |
| COG3250 | LacZ | 0.004 | 992 | 1094 | 50 | 164 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDF79916.1 | 0.0 | 6 | 1142 | 3 | 1155 |
| AUS07342.1 | 0.0 | 27 | 1142 | 5 | 1130 |
| APY12820.1 | 0.0 | 22 | 1145 | 14 | 1145 |
| ANW95880.1 | 0.0 | 27 | 1142 | 47 | 1172 |
| AWW28864.1 | 0.0 | 27 | 1149 | 31 | 1164 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 1.80e-104 | 27 | 1140 | 42 | 1136 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 2.45e-81 | 20 | 1093 | 24 | 1072 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 6.10e-81 | 20 | 1093 | 24 | 1072 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 0.0 | 6 | 1142 | 3 | 1155 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
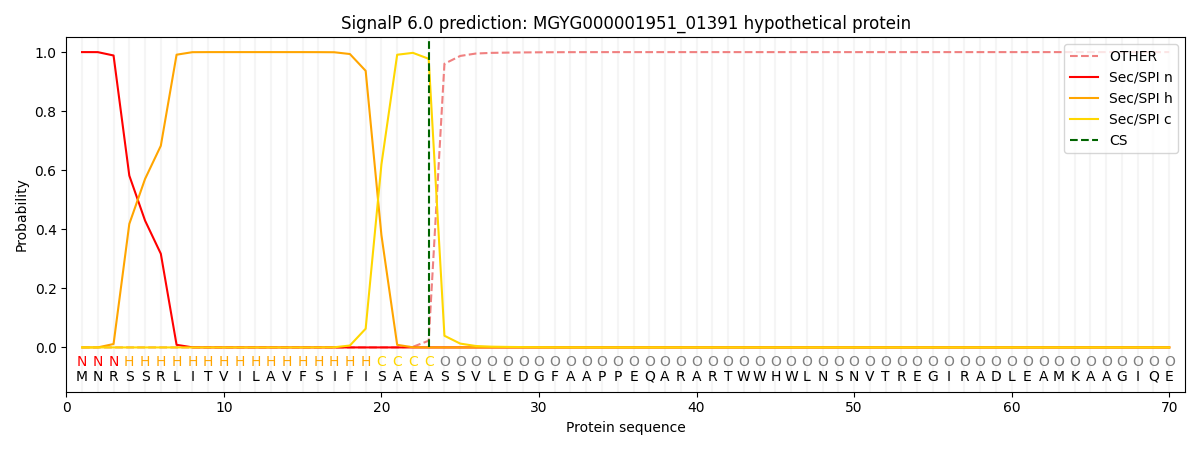
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000237 | 0.999160 | 0.000149 | 0.000149 | 0.000141 | 0.000130 |