You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001955_00422
You are here: Home > Sequence: MGYG000001955_00422
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Frisingicoccus sp900549105 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Frisingicoccus; Frisingicoccus sp900549105 | |||||||||||
| CAZyme ID | MGYG000001955_00422 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 70846; End: 72342 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 91 | 332 | 1.7e-94 | 0.9915611814345991 |
| CBM2 | 407 | 490 | 6.8e-17 | 0.7722772277227723 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.65e-67 | 89 | 340 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| smart00637 | CBD_II | 2.35e-12 | 419 | 488 | 8 | 76 | CBD_II domain. |
| COG2730 | BglC | 2.66e-09 | 144 | 341 | 102 | 280 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam00553 | CBM_2 | 3.95e-09 | 419 | 493 | 15 | 88 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| pfam09478 | CBM49 | 2.97e-05 | 402 | 479 | 2 | 72 | Carbohydrate binding domain CBM49. This domain is found at the C terminal of cellulases and in vitro binding studies have shown it to binds to crystalline cellulose. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFA47670.1 | 4.57e-140 | 79 | 487 | 203 | 623 |
| BCA89229.1 | 1.28e-139 | 74 | 497 | 85 | 512 |
| BCS56678.1 | 1.03e-138 | 74 | 497 | 85 | 512 |
| BAN76511.1 | 2.34e-137 | 74 | 497 | 85 | 512 |
| BCV18962.1 | 8.84e-133 | 74 | 372 | 61 | 358 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 3.67e-93 | 79 | 372 | 11 | 300 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 4XZB_A | 1.45e-86 | 79 | 368 | 10 | 299 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 3PZT_A | 3.44e-85 | 79 | 368 | 35 | 320 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 3.47e-84 | 79 | 368 | 10 | 298 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 1A3H_A | 6.26e-81 | 79 | 372 | 7 | 297 | EndoglucanaseCel5a From Bacillus Agaradherans At 1.6a Resolution [Salipaludibacillus agaradhaerens],2A3H_A Cellobiose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 2.0 A Resolution [Salipaludibacillus agaradhaerens],3A3H_A Cellotriose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 1.6 A Resolution [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q07940 | 2.39e-96 | 83 | 366 | 8 | 288 | Endoglucanase 4 OS=Ruminococcus albus OX=1264 GN=Eg IV PE=1 SV=1 |
| P22541 | 4.28e-96 | 63 | 382 | 92 | 417 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| P07983 | 3.92e-86 | 79 | 415 | 40 | 380 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P10475 | 8.46e-85 | 79 | 409 | 40 | 371 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P15704 | 6.92e-83 | 79 | 415 | 45 | 379 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
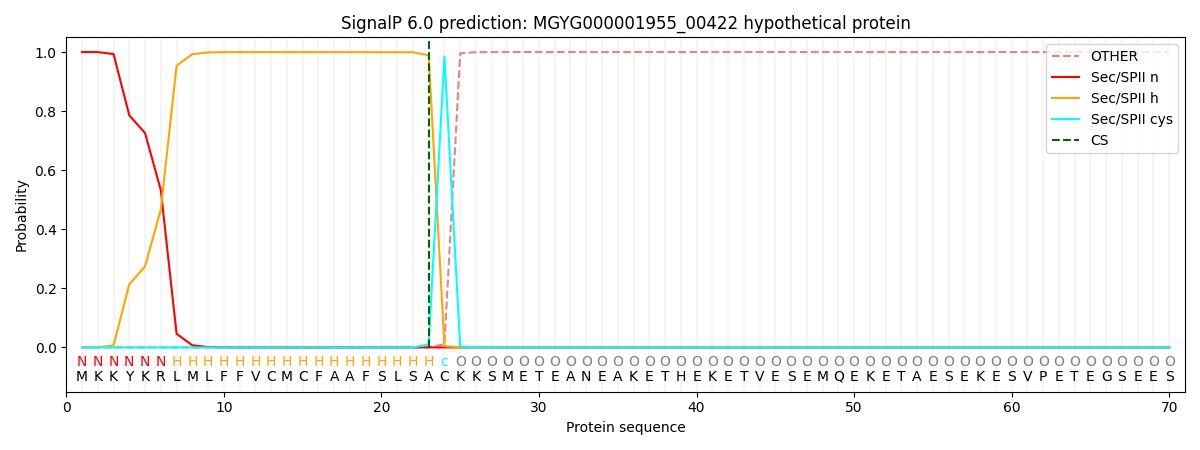
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000007 | 1.000066 | 0.000000 | 0.000000 | 0.000000 |

