You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001965_00356
You are here: Home > Sequence: MGYG000001965_00356
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-510 sp900550475 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-510; CAG-510 sp900550475 | |||||||||||
| CAZyme ID | MGYG000001965_00356 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 109743; End: 112871 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 621 | 852 | 1.1e-53 | 0.9327354260089686 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16192 | PMT_4TMC | 4.20e-27 | 867 | 1036 | 7 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG1928 | PMT1 | 1.13e-21 | 629 | 1041 | 50 | 696 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG5542 | COG5542 | 3.01e-16 | 64 | 364 | 71 | 362 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| pfam02366 | PMT | 4.68e-13 | 621 | 789 | 15 | 197 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This is a family of Dolichyl-phosphate-mannose-protein mannosyltransferase proteins EC:2.4.1.109. These proteins are responsible for O-linked glycosylation of proteins, they catalyze the reaction:- Dolichyl phosphate D-mannose + protein <=> dolichyl phosphate + O-D-mannosyl-protein. Also in this family is Drosophila rotated abdomen protein which is a putative mannosyltransferase. This family appears to be distantly related to pfam02516 (A Bateman pers. obs.). This family also contains sequences from ArnTs (4-amino-4-deoxy-L-arabinose lipid A transferase). They catalyze the addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N) to the lipid A moiety of the lipopolysaccharide. This is a critical modification enabling bacteria (e.g. Escherichia coli and Salmonella typhimurium) to resist killing by antimicrobial peptides such as polymyxins. Members such as undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase are predicted to have 12 trans-membrane regions. The N-terminal portion of these proteins is hypothesized to have a conserved glycosylation activity which is shared between distantly related oligosaccharyltransferases ArnT and PglB families. |
| COG4346 | COG4346 | 1.43e-12 | 643 | 989 | 127 | 414 | Predicted membrane-bound dolichyl-phosphate-mannose-protein mannosyltransferase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM01664.1 | 0.0 | 34 | 1042 | 34 | 1087 |
| BCN30459.1 | 8.92e-310 | 32 | 1042 | 38 | 1060 |
| BBF42044.1 | 8.10e-269 | 133 | 1041 | 2 | 921 |
| AIQ15817.1 | 2.92e-199 | 34 | 1039 | 10 | 1030 |
| AIQ66842.1 | 1.17e-194 | 34 | 1039 | 10 | 1030 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WN04 | 1.30e-29 | 629 | 1038 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 1.30e-29 | 629 | 1038 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| L8F4Z2 | 2.27e-27 | 629 | 1038 | 67 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| Q8NRZ6 | 2.40e-26 | 652 | 1038 | 99 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
| Q2LZ62 | 1.12e-08 | 850 | 1041 | 685 | 900 | Protein O-mannosyltransferase 1 OS=Drosophila pseudoobscura pseudoobscura OX=46245 GN=rt PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000007 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
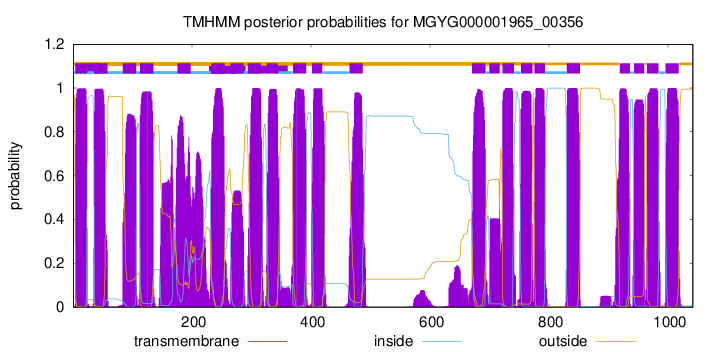
| start | end |
|---|---|
| 4 | 23 |
| 36 | 58 |
| 84 | 106 |
| 113 | 135 |
| 176 | 198 |
| 233 | 255 |
| 265 | 287 |
| 294 | 316 |
| 326 | 344 |
| 370 | 392 |
| 402 | 419 |
| 465 | 487 |
| 671 | 693 |
| 700 | 717 |
| 722 | 741 |
| 753 | 772 |
| 776 | 793 |
| 830 | 852 |
| 919 | 936 |
| 943 | 960 |
| 965 | 984 |
| 996 | 1018 |
