You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001972_01333
You are here: Home > Sequence: MGYG000001972_01333
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900315955 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900315955 | |||||||||||
| CAZyme ID | MGYG000001972_01333 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8410; End: 11250 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 136 | 440 | 6.5e-83 | 0.9855072463768116 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.47e-42 | 127 | 443 | 7 | 271 | Cellulase (glycosyl hydrolase family 5). |
| sd00036 | LRR_3 | 5.49e-20 | 743 | 850 | 6 | 117 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| pfam13306 | LRR_5 | 5.56e-20 | 741 | 842 | 2 | 127 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
| sd00036 | LRR_3 | 9.92e-19 | 740 | 844 | 26 | 134 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| pfam13306 | LRR_5 | 3.81e-18 | 708 | 816 | 12 | 122 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT65310.1 | 4.87e-88 | 81 | 455 | 117 | 506 |
| AUS04354.1 | 1.67e-80 | 34 | 470 | 72 | 502 |
| QSB27529.1 | 7.51e-80 | 66 | 470 | 110 | 497 |
| QGK76460.1 | 1.04e-77 | 99 | 464 | 143 | 492 |
| QMI80089.1 | 1.28e-77 | 101 | 470 | 219 | 564 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OYC_A | 3.55e-78 | 85 | 470 | 19 | 389 | GH5endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYC_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107] |
| 6HA9_A | 2.53e-77 | 85 | 470 | 19 | 389 | Structureof an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HA9_B Structure of an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_A Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_B Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107] |
| 4W8A_A | 2.57e-76 | 109 | 476 | 3 | 373 | Crystalstructure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form [uncultured bacterium],4W8B_A Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in complex with XXLG [uncultured bacterium] |
| 4NF7_A | 1.32e-49 | 104 | 450 | 3 | 336 | Crystalstructure of the GH5 family catalytic domain of Endo-1,4-beta-glucanase Cel5C from Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316] |
| 6Q1I_A | 5.35e-49 | 109 | 457 | 14 | 336 | GH5-4broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum],6Q1I_B GH5-4 broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54937 | 6.32e-48 | 109 | 487 | 39 | 394 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P20847 | 3.65e-47 | 104 | 450 | 34 | 367 | Endoglucanase 1 OS=Butyrivibrio fibrisolvens OX=831 GN=end1 PE=3 SV=1 |
| P28623 | 1.06e-45 | 82 | 449 | 18 | 350 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| A7LXT7 | 3.78e-45 | 111 | 441 | 152 | 475 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02653 PE=1 SV=1 |
| P28621 | 9.35e-45 | 111 | 449 | 44 | 352 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
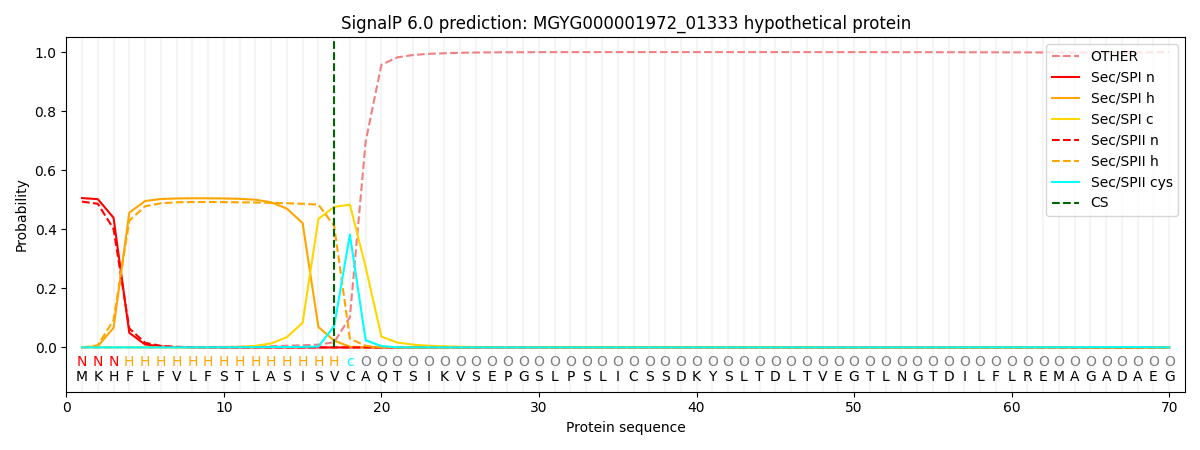
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001265 | 0.498354 | 0.499685 | 0.000326 | 0.000174 | 0.000169 |
