You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001972_01829
You are here: Home > Sequence: MGYG000001972_01829
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
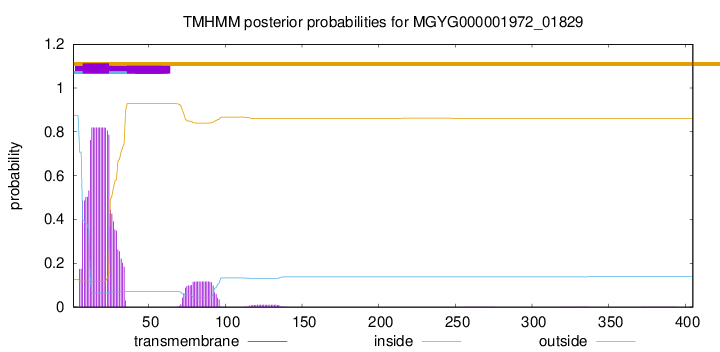
TMHMM annotations
Basic Information help
| Species | Prevotella sp900315955 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900315955 | |||||||||||
| CAZyme ID | MGYG000001972_01829 | |||||||||||
| CAZy Family | GH88 | |||||||||||
| CAZyme Description | Unsaturated chondroitin disaccharide hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3420; End: 4637 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH88 | 76 | 399 | 8.2e-123 | 0.9787234042553191 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07470 | Glyco_hydro_88 | 7.22e-10 | 64 | 288 | 1 | 225 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| cd00249 | AGE | 0.004 | 208 | 317 | 74 | 171 | AGE domain; N-acyl-D-glucosamine 2-epimerase domain; Responsible for intermediate epimerization during biosynthesis of N-acetylneuraminic acid. Catalytic mechanism is believed to be via nucleotide elimination and readdition and is ATP modulated. AGE is structurally and mechanistically distinct from the other four types of epimerases. The AGE domain monomer is composed of an alpha(6)/alpha(6)-barrel, the structure of which is also found in glucoamylase and cellulase. The active form is a homodimer. The alignment also contains subtype III mannose 6-phosphate isomerases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEH14502.1 | 1.29e-219 | 6 | 400 | 1 | 396 |
| AGB28567.1 | 2.21e-187 | 38 | 399 | 31 | 391 |
| QIU93750.1 | 1.03e-186 | 37 | 400 | 31 | 394 |
| QGT72848.1 | 2.08e-186 | 37 | 400 | 31 | 394 |
| QDM10821.1 | 4.19e-186 | 37 | 400 | 31 | 394 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WIW_A | 2.52e-98 | 62 | 399 | 56 | 387 | Crystalstructure of unsaturated glucuronyl hydrolase specific for heparin [Pedobacter heparinus DSM 2366] |
| 2ZZR_A | 2.47e-49 | 39 | 402 | 32 | 391 | Crystalstructure of unsaturated glucuronyl hydrolase from Streptcoccus agalactiae [Streptococcus agalactiae] |
| 3ANJ_A | 2.52e-49 | 39 | 402 | 33 | 392 | Crystalstructure of unsaturated glucuronyl hydrolase from Streptcoccus agalactiae [Streptococcus agalactiae serogroup III] |
| 3WUX_A | 3.53e-49 | 39 | 402 | 33 | 392 | Crystalstructure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae [Streptococcus agalactiae NEM316] |
| 3ANI_A | 1.35e-48 | 39 | 402 | 33 | 392 | Crystalstructure of unsaturated glucuronyl hydrolase mutant D175N from Streptcoccus agalactiae [Streptococcus agalactiae serogroup III],3ANK_A Crystal structure of unsaturated glucuronyl hydrolase mutant D175N from Streptcoccus agalactiae complexed with dGlcA-GalNAc6S [Streptococcus agalactiae serogroup III] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KLZ3 | 5.07e-100 | 74 | 400 | 72 | 394 | Unsaturated glucuronyl hydrolase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21900 PE=1 SV=1 |
| Q8E372 | 1.38e-48 | 39 | 402 | 33 | 392 | Unsaturated chondroitin disaccharide hydrolase OS=Streptococcus agalactiae serotype III (strain NEM316) OX=211110 GN=gbs1889 PE=1 SV=1 |
| Q9A0T3 | 1.10e-46 | 39 | 402 | 34 | 393 | Unsaturated chondroitin disaccharide hydrolase OS=Streptococcus pyogenes serotype M1 OX=301447 GN=ugl PE=1 SV=1 |
| Q9RC92 | 2.55e-46 | 60 | 403 | 23 | 367 | Unsaturated glucuronyl hydrolase OS=Bacillus sp. (strain GL1) OX=84635 GN=ugl PE=1 SV=1 |
| Q8DR77 | 1.06e-45 | 39 | 402 | 31 | 390 | Unsaturated chondroitin disaccharide hydrolase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=ugl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
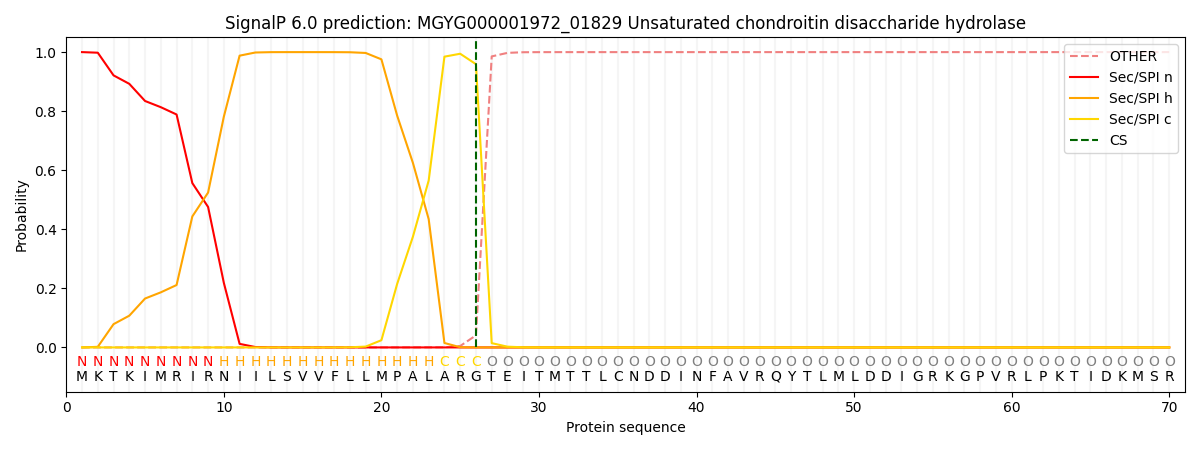
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000301 | 0.998993 | 0.000223 | 0.000152 | 0.000147 | 0.000145 |