You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001975_00819
You are here: Home > Sequence: MGYG000001975_00819
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900552515 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900552515 | |||||||||||
| CAZyme ID | MGYG000001975_00819 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | Keratan-sulfate endo-1,4-beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17787; End: 18566 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 28 | 255 | 2.5e-56 | 0.9956521739130435 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08023 | GH16_laminarinase_like | 7.38e-73 | 28 | 255 | 1 | 235 | Laminarinase, member of the glycosyl hydrolase family 16. Laminarinase, also known as glucan endo-1,3-beta-D-glucosidase, is a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
| cd00413 | Glyco_hydrolase_16 | 4.13e-28 | 30 | 255 | 1 | 210 | glycosyl hydrolase family 16. The O-Glycosyl hydrolases are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A glycosyl hydrolase classification system based on sequence similarity has led to the definition of more than 95 different families inlcuding glycosyl hydrolase family 16. Family 16 includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
| COG2273 | BglS | 2.08e-22 | 9 | 256 | 23 | 264 | Beta-glucanase, GH16 family [Carbohydrate transport and metabolism]. |
| cd08024 | GH16_CCF | 5.58e-21 | 26 | 227 | 1 | 271 | Coelomic cytolytic factor, member of glycosyl hydrolase family 16. Subgroup of glucanases of unknown function that are related to beta-GRP (beta-1,3-glucan recognition protein), but contain active site residues. Beta-GRPs are one group of pattern recognition receptors (PRRs), also referred to as biosensor proteins, that complexes with pathogen-associated beta-1,3-glucans and then transduces signals necessary for activation of an appropriate innate immune response. Beta-GRPs are present in insects and lack all catalytic residues. This subgroup contains related proteins that still contain the active site and are widely distributed in eukaryotes. Their structures adopt a jelly roll fold with a deep active site channel harboring the catalytic residues, like those of other glycosyl hydrolase family 16 members. |
| pfam00722 | Glyco_hydro_16 | 8.21e-18 | 98 | 253 | 28 | 168 | Glycosyl hydrolases family 16. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO25177.1 | 6.67e-114 | 2 | 258 | 5 | 259 |
| AQX06254.1 | 1.39e-101 | 3 | 258 | 15 | 270 |
| SQG08214.1 | 1.39e-101 | 3 | 258 | 15 | 270 |
| AQX48301.1 | 1.39e-101 | 3 | 258 | 15 | 270 |
| QCQ43864.1 | 1.51e-101 | 20 | 258 | 26 | 263 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6T2R_AAA | 2.36e-97 | 20 | 258 | 26 | 264 | ChainAAA, Beta-glycosidase [Bacteroides caccae] |
| 5WUT_A | 2.42e-30 | 25 | 256 | 6 | 235 | Crystalstructure of laminarinase from Flavobacterium sp. UMI-01 [Flavobacterium sp.],5WUT_B Crystal structure of laminarinase from Flavobacterium sp. UMI-01 [Flavobacterium sp.] |
| 6XOF_A | 3.94e-29 | 21 | 256 | 21 | 267 | ChainA, GH16 family protein [uncultured bacterium] |
| 6XQF_A | 2.11e-28 | 21 | 256 | 21 | 267 | ChainA, GH16 family protein [uncultured bacterium],6XQG_A Chain A, GH16 family protein [uncultured bacterium],6XQH_A Chain A, GH16 family protein [uncultured bacterium],6XQL_A Chain A, GH16 family protein [uncultured bacterium],6XQM_A Chain A, GH16 family protein [uncultured bacterium] |
| 3ILN_A | 5.51e-28 | 21 | 255 | 3 | 247 | ChainA, Laminarinase [Rhodothermus marinus],3ILN_B Chain B, Laminarinase [Rhodothermus marinus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ZG90 | 5.59e-43 | 12 | 257 | 46 | 289 | Keratan-sulfate endo-1,4-beta-galactosidase OS=Sphingobacterium multivorum OX=28454 PE=1 SV=1 |
| P45798 | 1.12e-28 | 21 | 255 | 38 | 282 | Beta-glucanase OS=Rhodothermus marinus OX=29549 GN=bglA PE=1 SV=1 |
| P23903 | 2.31e-26 | 26 | 255 | 425 | 679 | Glucan endo-1,3-beta-glucosidase A1 OS=Niallia circulans OX=1397 GN=glcA PE=1 SV=1 |
| Q27082 | 3.84e-20 | 1 | 256 | 1 | 253 | Clotting factor G alpha subunit OS=Tachypleus tridentatus OX=6853 PE=1 SV=1 |
| Q9Z3Q2 | 1.69e-12 | 64 | 257 | 276 | 459 | Endo-1,3-1,4-beta-glycanase EglC OS=Rhizobium meliloti (strain 1021) OX=266834 GN=eglC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
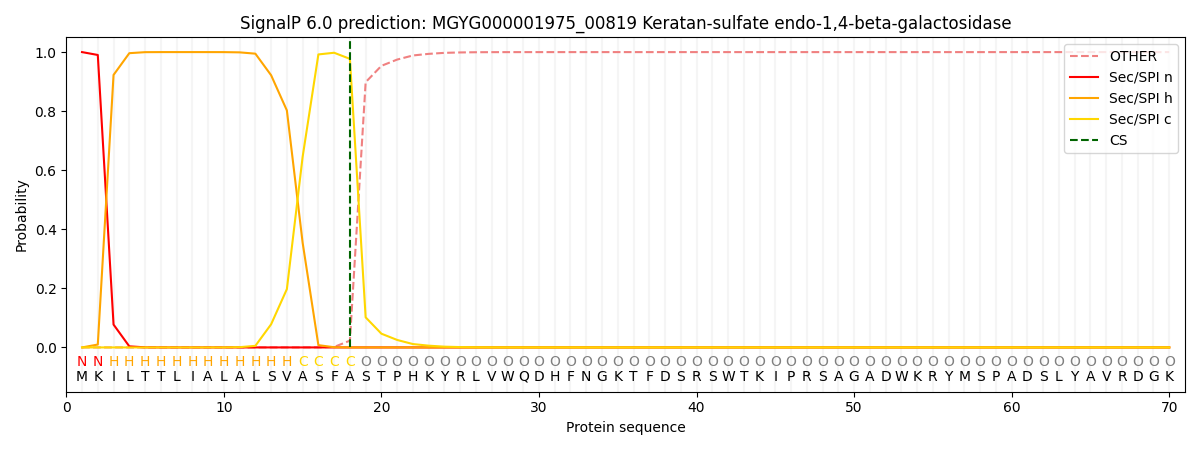
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000263 | 0.999105 | 0.000172 | 0.000166 | 0.000149 | 0.000145 |
