You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001977_00496
You are here: Home > Sequence: MGYG000001977_00496
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000001977_00496 | |||||||||||
| CAZy Family | GH97 | |||||||||||
| CAZyme Description | Glucan 1,4-alpha-glucosidase SusB | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 39168; End: 41384 Strand: + | |||||||||||
Full Sequence Download help
| MKLKKNLSLI AFLCISFIAN AQQKLSSPDG NLEMTFTLDG QGTPTYELTY KQKEVIKPSR | 60 |
| LGLELKKEDA NAKTDFEWTD KKDIDKLDIK TNLYNGFELK DVQTSTNDET WQPVWGEEKE | 120 |
| IRNHYNELAV TLNQPANDRF IIIRFRLFND GLGFRYEFPQ QKNLNYFVIK EEHSQFAMAG | 180 |
| DHTAYWIPGD YDTQEYDYTV SRLSEIRGLF SKAYTENSSQ TAFSPTGVQT ALMMKTDDGL | 240 |
| YINLHEAALI DYACMHLNLD DKNMIFESWL TPDAQGDKGY MQTPCTSPWR TVIVSDDARN | 300 |
| ILASRITLNL NEPCKIADTS WIKPVKYIGV WWDMITGKGS WAYTDDLSSV KLGETDYNKT | 360 |
| KPNGKHSANT ANVKRYIDFA AEHGFDQVLV EGWNEGWEDW FGKSKDYVFD FVTPYPDFNV | 420 |
| KELQSYAASK GVKLMMHHET SASVRNYERH MDQAYQFMAD NGYNSVKSGY VGNIIPRGEH | 480 |
| HYGQWMDNHY LYAVKKAADY KIMVNAHEAV RPTGICRTYP NLIGNESARG TEYESFGGNN | 540 |
| VNHTTILPFT RLIGGPMDYT PGIFETRCNK MNPTNNSQVR STLARQLALY VTMYSPLQMA | 600 |
| ADIPENYERF MDAFQFIKDV AIDWDETKYL EAEPGDYITI ARKAKGTNDW YIGCTADENG | 660 |
| HSSKLIFDFL DPDKKYIATV YADAKDADWK DNPQAYTIRK GIVTNKSKLN LHAASGGGYA | 720 |
| ISIKEVKDKS ELKGIKRL | 738 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH97 | 12 | 723 | 2.3e-258 | 0.9920760697305864 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam10566 | Glyco_hydro_97 | 5.23e-148 | 318 | 621 | 1 | 278 | Glycoside hydrolase 97. This domain is the catalytic region of the bacterial glycosyl-hydrolase family 97. This central part of the GH97 family protein sequences represents a typical and complete (beta/alpha)8-barrel or catalytic TIM-barrel type domain. The N- and C-terminal parts of the sequences, mainly consisting of beta-strands, form two additional non-catalytic domains. In all known glycosidases with the (beta-alpha)8-barrel fold, the amino acid residues at the active site are located on the C-termini of the beta-strands. |
| pfam14508 | GH97_N | 7.34e-103 | 27 | 313 | 1 | 235 | Glycosyl-hydrolase 97 N-terminal. This N-terminal domain of glycosyl-hydrolase-97 contributes part of the active site pocket. It is also important for contact with the catalytic and C-terminal domains of the whole. |
| pfam14509 | GH97_C | 1.86e-45 | 624 | 724 | 1 | 97 | Glycosyl-hydrolase 97 C-terminal, oligomerization. Glycosyl-hydrolase-97 is made up of three tightly linked and highly conserved globular domains. The C-terminal domain is found to be necessary for oligomerization of the whole molecule in order to create the active-site pocket and the Ca++-binding site. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALA72849.1 | 0.0 | 1 | 738 | 1 | 738 |
| QJR61692.1 | 0.0 | 1 | 738 | 1 | 738 |
| AII67018.1 | 0.0 | 1 | 738 | 1 | 738 |
| QJR53512.1 | 0.0 | 1 | 738 | 1 | 738 |
| AND18771.1 | 0.0 | 1 | 738 | 1 | 738 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2D73_A | 0.0 | 19 | 738 | 19 | 738 | CrystalStructure Analysis of SusB [Bacteroides thetaiotaomicron VPI-5482],2D73_B Crystal Structure Analysis of SusB [Bacteroides thetaiotaomicron VPI-5482],2ZQ0_A Crystal structure of SusB complexed with acarbose [Bacteroides thetaiotaomicron],2ZQ0_B Crystal structure of SusB complexed with acarbose [Bacteroides thetaiotaomicron] |
| 2JKA_A | 0.0 | 24 | 738 | 13 | 727 | Nativestructure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],2JKA_B Native structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],2JKE_A Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with deoxynojirimycin [Bacteroides thetaiotaomicron VPI-5482],2JKE_B Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with deoxynojirimycin [Bacteroides thetaiotaomicron VPI-5482],2JKP_A Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with castanospermine [Bacteroides thetaiotaomicron VPI-5482],2JKP_B Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with castanospermine [Bacteroides thetaiotaomicron VPI-5482] |
| 3WFA_A | 0.0 | 22 | 738 | 2 | 718 | Catalyticrole of the calcium ion in GH97 inverting glycoside hydrolase [Bacteroides thetaiotaomicron],3WFA_B Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase [Bacteroides thetaiotaomicron] |
| 5HQ4_A | 1.18e-170 | 25 | 724 | 3 | 659 | AGlycoside Hydrolase Family 97 enzyme from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8],5HQA_A A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8] |
| 5HQC_A | 1.18e-170 | 25 | 724 | 3 | 659 | AGlycoside Hydrolase Family 97 enzyme R171K variant from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G8JZS4 | 0.0 | 1 | 738 | 1 | 738 | Glucan 1,4-alpha-glucosidase SusB OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=susB PE=1 SV=1 |
| Q8A6L0 | 2.28e-67 | 9 | 724 | 8 | 662 | Retaining alpha-galactosidase OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_1871 PE=1 SV=1 |
| D7CFN7 | 6.75e-30 | 26 | 720 | 44 | 617 | Probable retaining alpha-galactosidase OS=Streptomyces bingchenggensis (strain BCW-1) OX=749414 GN=SBI_01652 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
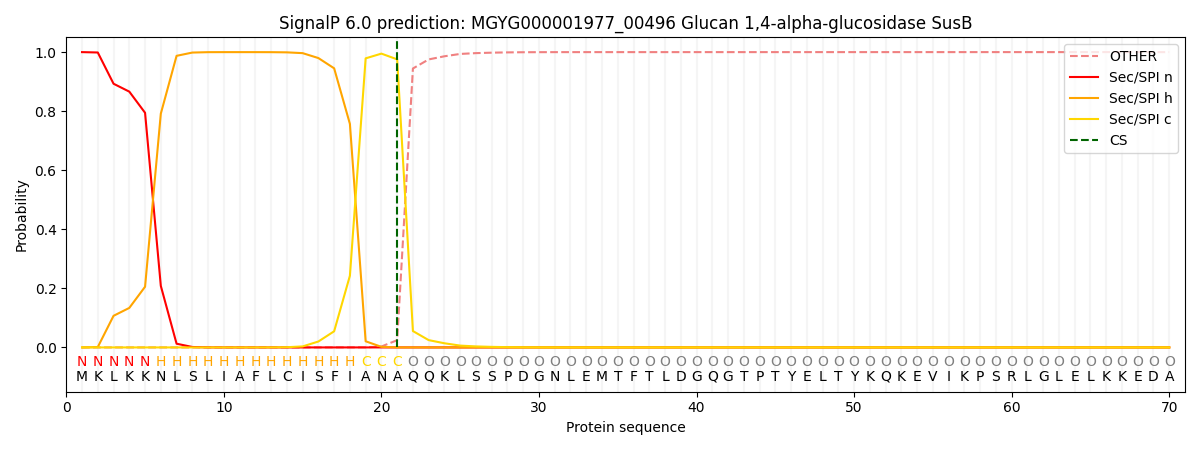
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000414 | 0.998603 | 0.000378 | 0.000190 | 0.000200 | 0.000180 |
