You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001977_01532
You are here: Home > Sequence: MGYG000001977_01532
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000001977_01532 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14546; End: 15979 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 91 | 250 | 9.6e-28 | 0.39211136890951276 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01229 | Glyco_hydro_39 | 6.58e-09 | 54 | 265 | 21 | 263 | Glycosyl hydrolases family 39. |
| pfam02449 | Glyco_hydro_42 | 6.86e-05 | 79 | 174 | 19 | 138 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| pfam00150 | Cellulase | 1.34e-04 | 62 | 205 | 21 | 163 | Cellulase (glycosyl hydrolase family 5). |
| pfam00331 | Glyco_hydro_10 | 3.29e-04 | 88 | 204 | 46 | 164 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO70685.1 | 6.09e-166 | 34 | 477 | 38 | 480 |
| AEI51083.1 | 1.93e-155 | 33 | 477 | 41 | 483 |
| AEI51085.1 | 6.66e-149 | 38 | 477 | 42 | 498 |
| AQG80712.1 | 1.22e-145 | 34 | 476 | 55 | 514 |
| CCH00422.1 | 3.43e-142 | 37 | 476 | 48 | 490 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
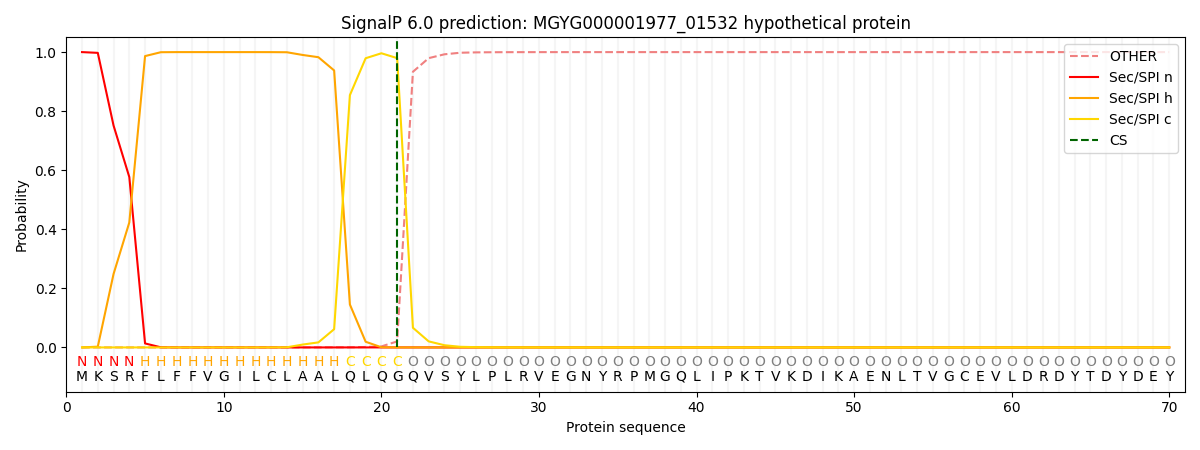
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000192 | 0.999249 | 0.000145 | 0.000136 | 0.000125 | 0.000124 |
