You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001994_00300
You are here: Home > Sequence: MGYG000001994_00300
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
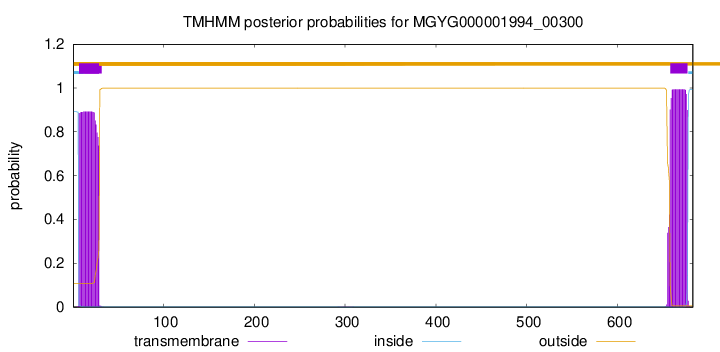
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Schaedlerella; | |||||||||||
| CAZyme ID | MGYG000001994_00300 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8060; End: 10111 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 3.25e-20 | 228 | 405 | 57 | 237 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| PTZ00449 | PTZ00449 | 7.33e-20 | 413 | 506 | 573 | 666 | 104 kDa microneme/rhoptry antigen; Provisional |
| PTZ00449 | PTZ00449 | 1.25e-19 | 413 | 503 | 579 | 669 | 104 kDa microneme/rhoptry antigen; Provisional |
| PTZ00449 | PTZ00449 | 1.89e-18 | 416 | 506 | 579 | 669 | 104 kDa microneme/rhoptry antigen; Provisional |
| PTZ00449 | PTZ00449 | 7.65e-18 | 412 | 501 | 584 | 673 | 104 kDa microneme/rhoptry antigen; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEK17141.1 | 3.04e-111 | 1 | 406 | 1 | 398 |
| QOV20091.1 | 7.61e-107 | 39 | 405 | 28 | 387 |
| AWY97555.1 | 6.99e-95 | 27 | 406 | 22 | 391 |
| QNM11023.1 | 1.66e-85 | 4 | 406 | 4 | 448 |
| BCT44532.1 | 2.57e-83 | 69 | 406 | 126 | 452 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 3.59e-36 | 130 | 405 | 57 | 290 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 5.44e-34 | 130 | 405 | 425 | 658 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 6.98e-34 | 130 | 405 | 469 | 702 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
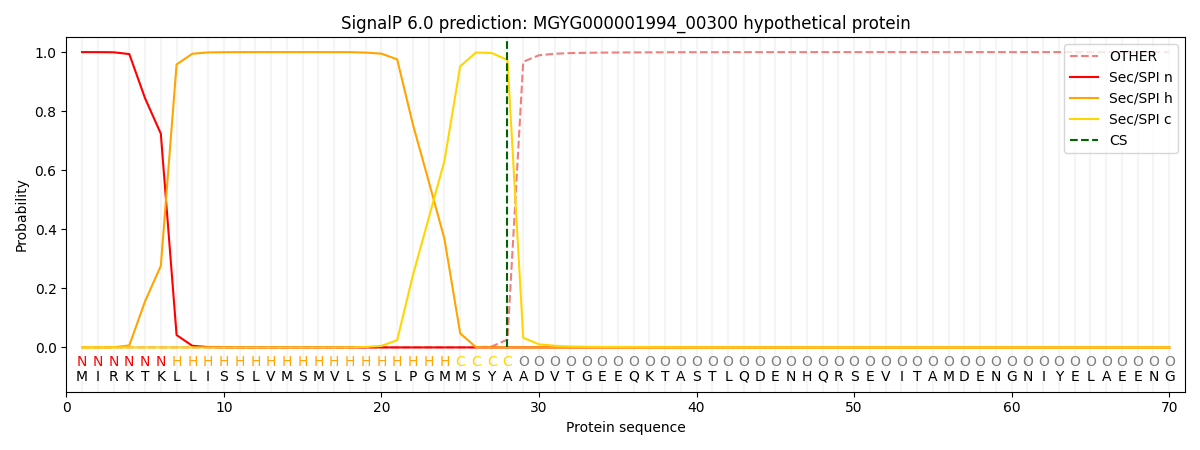
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000281 | 0.998914 | 0.000219 | 0.000244 | 0.000175 | 0.000148 |