You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001995_00709
You are here: Home > Sequence: MGYG000001995_00709
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900541965 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900541965 | |||||||||||
| CAZyme ID | MGYG000001995_00709 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Exo-beta-D-glucosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 106815; End: 109469 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 63 | 774 | 4.6e-83 | 0.6901595744680851 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.28e-38 | 102 | 534 | 53 | 442 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam18368 | Ig_GlcNase | 2.53e-27 | 770 | 874 | 1 | 102 | Exo-beta-D-glucosaminidase Ig-fold domain. This domain can be found in 2 glycoside hydrolase subfamily of beta-glucosaminidases (EC:3.2.1.165) such as CsxA, from Amycolatopsis orientalis that has exo-beta-D-glucosaminidase (exo-chitosanase) activity. It has an immunoglobulin-like topology. |
| PRK10150 | PRK10150 | 9.02e-17 | 97 | 529 | 52 | 460 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 8.96e-15 | 117 | 496 | 115 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 5.68e-11 | 254 | 353 | 9 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUR48650.1 | 0.0 | 1 | 876 | 1 | 874 |
| QCY54664.1 | 0.0 | 1 | 876 | 1 | 874 |
| QUT20722.1 | 0.0 | 1 | 876 | 1 | 874 |
| QKH99571.1 | 0.0 | 1 | 876 | 1 | 874 |
| AST56014.1 | 0.0 | 1 | 876 | 1 | 874 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VZS_A | 7.75e-74 | 66 | 874 | 78 | 889 | ChitosanProduct complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZS_B Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2X05_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X05_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis] |
| 2VZO_A | 1.06e-73 | 66 | 874 | 78 | 889 | Crystalstructure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZO_B Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis] |
| 2VZT_A | 4.96e-73 | 66 | 874 | 78 | 889 | Complexof Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZT_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZV_A Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis],2VZV_B Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis] |
| 2VZU_A | 2.32e-72 | 66 | 874 | 78 | 889 | Complexof Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZU_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis] |
| 5N6U_A | 1.10e-51 | 69 | 880 | 43 | 834 | Crystalstructure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q75W54 | 2.51e-190 | 37 | 883 | 6 | 944 | Mannosylglycoprotein endo-beta-mannosidase OS=Arabidopsis thaliana OX=3702 GN=EBM PE=1 SV=3 |
| Q5H7P5 | 4.40e-187 | 37 | 858 | 3 | 928 | Mannosylglycoprotein endo-beta-mannosidase OS=Lilium longiflorum OX=4690 GN=EBM PE=1 SV=4 |
| D4AUH1 | 8.97e-82 | 30 | 878 | 28 | 879 | Exo-beta-D-glucosaminidase ARB_07888 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07888 PE=1 SV=1 |
| Q4R1C4 | 1.41e-76 | 35 | 856 | 30 | 868 | Exo-beta-D-glucosaminidase OS=Hypocrea jecorina OX=51453 GN=gls93 PE=1 SV=1 |
| C0LRA7 | 1.72e-75 | 22 | 856 | 15 | 866 | Exo-beta-D-glucosaminidase OS=Hypocrea virens OX=29875 GN=gls1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
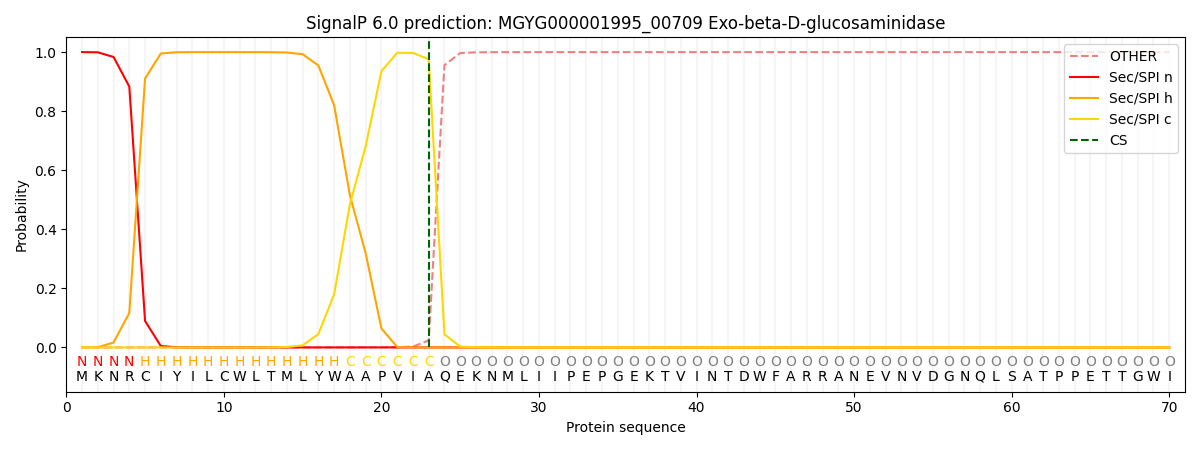
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000313 | 0.999028 | 0.000207 | 0.000157 | 0.000147 | 0.000138 |

