You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001995_01020
You are here: Home > Sequence: MGYG000001995_01020
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
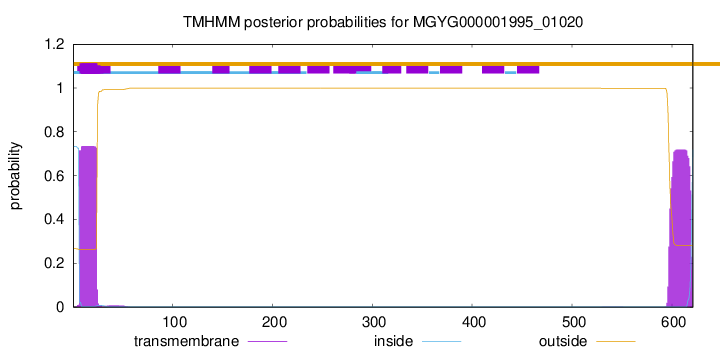
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900541965 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900541965 | |||||||||||
| CAZyme ID | MGYG000001995_01020 | |||||||||||
| CAZy Family | GH116 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 63587; End: 65452 Strand: - | |||||||||||
Full Sequence Download help
| MNSMKTHSLF FLLFCWVICH SMVAQKRQVA DGYPIANFAL TVNASVSGDF FGQIMQNPGT | 60 |
| LGDWLTNRGA VIFSIEKDGK RQLFSEFSKK NIIREFPFVK NDYKGSSLTK VTFTTEAFCP | 120 |
| LGVNDAETSA LPVLMLEICC SNPTSKEEHF TLVAQPDENL AKDMSACEHD GYTGISNKDN | 180 |
| CQITTDQAET LWAGQRLSIP VSLQAGEKKK IKVLFTFRDE NWISSINFRT LDDLSTYAYK | 240 |
| MWPAFKDKTT AFGLAIPKTG DKELDTYVRW YMIPGISLTK WTKKGELVTM GYCELNQRDS | 300 |
| YWTSWLHLVF YKDLERKMIE ESIEWQQPSG KIPTTILPLI EREDDLDINA FFILRIARFY | 360 |
| RYYHNKKDLT KYWPAMKNAM NWLIYRDTDK IGLPAQVSFW GDWKDVKGVE GRKYSPFTCL | 420 |
| IYLSALKEMV YLAEECGDQT GGNYYRAAYQ KGYETVNKDV KKGGLWNGSY YCQIWKDGSV | 480 |
| NDKILQDQTI GILFDVVPRE RALKIIETLN TKSMTPYGVA ETFPYYDDSF GYPSATYHNG | 540 |
| GVWPWVSFMD CWARINMGRQ EEAIDIIKKV GRADLVNSGD WSPNEHINSL TGENLGYHIQ | 600 |
| GWNTGLFGLV YFGLVNIGIM H | 621 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH116 | 335 | 573 | 1.7e-22 | 0.6556473829201102 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam04685 | DUF608 | 9.23e-18 | 347 | 570 | 99 | 324 | Glycosyl-hydrolase family 116, catalytic region. This represents a family of archaeal, bacterial and eukaryotic glycosyl hydrolases, that belong to superfamily GH116. The primary catabolic pathway for glucosylceramide is catalysis by the lysosomal enzyme glucocerebrosidase. In higher eukaryotes, glucosylceramide is the precursor of glycosphingolipids, a complex group of ubiquitous membrane lipids. Mutations in the human protein cause motor-neurone defects in hereditary spastic paraplegia. The catalytic nucleophile, identified in UniProtKB:Q97YG8_SULSO, is a glutamine-335, with the likely acid/base at Asp-442 and the aspartates at Asp-406 and Asp-458 residues also playing a role in the catalysis of glucosides and xylosides that are beta-bound to hydrophobic groups. The family is defined as GH116, which presently includes enzymes with beta-glucosidase, EC:3.2.1.21, beta-xylosidase, EC:3.2.1.37, and glucocerebrosidase EC:3.2.1.45 activity. |
| COG4354 | COG4354 | 2.25e-15 | 347 | 593 | 442 | 697 | Uncharacterized protein, contains GBA2_N and DUF608 domains [Function unknown]. |
| COG3408 | GDB1 | 2.87e-11 | 307 | 545 | 308 | 539 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| pfam06202 | GDE_C | 1.09e-10 | 351 | 570 | 88 | 331 | Amylo-alpha-1,6-glucosidase. This family includes human glycogen branching enzyme AGL. This enzyme contains a number of distinct catalytic activities. It has been shown for the yeast homolog GDB1 that mutations in this region disrupt the enzymes Amylo-alpha-1,6-glucosidase (EC:3.2.1.33). |
| pfam17167 | Glyco_hydro_36 | 6.94e-07 | 401 | 616 | 188 | 419 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUR48358.1 | 0.0 | 1 | 621 | 1 | 620 |
| QCY58135.1 | 0.0 | 1 | 621 | 1 | 620 |
| QJE29784.1 | 0.0 | 1 | 621 | 1 | 620 |
| QUT52127.1 | 0.0 | 1 | 621 | 1 | 620 |
| ANH81218.1 | 1.24e-279 | 9 | 616 | 6 | 609 |
SignalP and Lipop Annotations help
This protein is predicted as SP
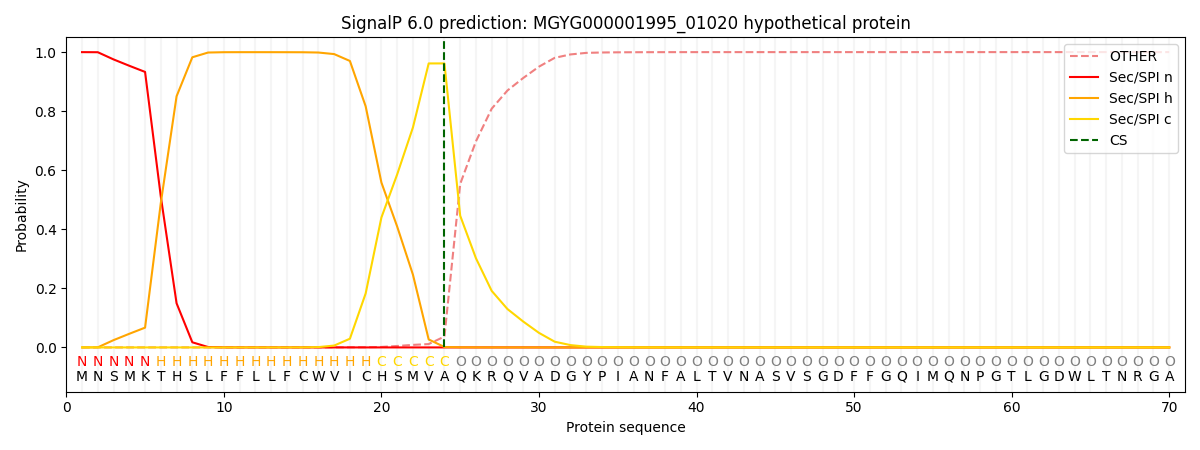
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000578 | 0.998394 | 0.000471 | 0.000170 | 0.000170 | 0.000178 |