You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001995_01157
You are here: Home > Sequence: MGYG000001995_01157
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
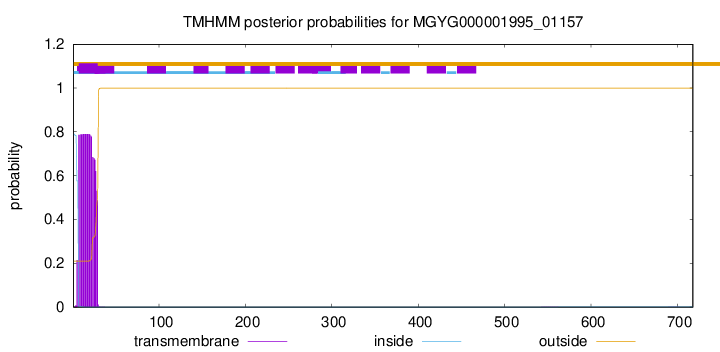
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900541965 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900541965 | |||||||||||
| CAZyme ID | MGYG000001995_01157 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | Alpha-galactosidase AgaA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30585; End: 32741 Strand: + | |||||||||||
Full Sequence Download help
| MKIRHILFVC MVAFTTVFHA SAKTIIPITT DNISLIYKVD EANGRLYQSY LGQKLSFSSD | 60 |
| IDQLPLGREA YLTHGMEDYF EPAIRVLHND GNPSLLLKYV SHETKQMQPG ANETVIILRD | 120 |
| EVYPVEVKLH FIAYPKENII KEYAEISHNE KKPVTLYNYA SSLLHLNSDK YYLTEFAGDW | 180 |
| AHEVNMRESE LSFGKKILDT KLGSRANMFC SPFFLLALDE KAEENSGEVL FGTIGWTGNY | 240 |
| RFTFEVDNLN GLRILSGINP YASEYTLKPG VVFRTPEFIF TYSSEGKGKA SRDFHRWARK | 300 |
| YQLKDGEKSR MTLLNNWEAT YFDFNEDKLV EIMGEAADLG VDMFLLDDGW FANKYPRSSD | 360 |
| HQGLGDWTET VTKLPNGIGK LVKEATDKGI KFGLWIEPEM VNPKSELYEK HMDWVIHLPN | 420 |
| RDEYYFRNQM VLDLSNPEVQ DYVYGVVDGL MTKYPGIAYF KWDCNSPITN IYSAYLKDKQ | 480 |
| SHLYIEYVRG LYSVLERIKA KYPNLPMMLC SGGGGRSDYE ALKYFTEFWP SDNTDPVERL | 540 |
| FIQWGFSHFF PVKSMAAHVT SWGKQPVKFR TDVAMMCKLG FDIRLHEMEP ADLQYCKEAV | 600 |
| ANFKRLSYVI LDGDQYRLKS PYESDHAAFL YANETQSKAV LFAFDIHPRY AEMLQPLRLQ | 660 |
| GLKKDARYEL KEINLVPGTK SRMPGTMTYS GEYLMKVGIP VFSSRHTVSH VIEITEVR | 718 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH36 | 27 | 700 | 1.2e-205 | 0.9941860465116279 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02065 | Melibiase | 1.65e-122 | 273 | 613 | 2 | 347 | Melibiase. Glycoside hydrolase families GH27, GH31 and GH36 form the glycoside hydrolase clan GH-D. Glycoside hydrolase family 36 can be split into 11 families, GH36A to GH36K. This family includes enzymes from GH36A-B and GH36D-K and from GH27. |
| cd14791 | GH36 | 1.29e-109 | 310 | 605 | 2 | 299 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| COG3345 | GalA | 7.72e-105 | 55 | 705 | 48 | 671 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
| pfam16875 | Glyco_hydro_36N | 1.30e-59 | 44 | 268 | 1 | 256 | Glycosyl hydrolase family 36 N-terminal domain. This domain is found at the N-terminus of many family 36 glycoside hydrolases. It has a beta-supersandwich fold. |
| pfam16874 | Glyco_hydro_36C | 1.82e-16 | 626 | 705 | 1 | 74 | Glycosyl hydrolase family 36 C-terminal domain. This domain is found at the C-terminus of many family 36 glycoside hydrolases. It has a beta-sandwich structure with a Greek key motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT48223.1 | 0.0 | 1 | 718 | 1 | 719 |
| QQA08975.1 | 0.0 | 1 | 714 | 1 | 715 |
| AII65838.1 | 0.0 | 1 | 714 | 1 | 715 |
| QMW85283.1 | 0.0 | 1 | 714 | 4 | 718 |
| AAO77957.1 | 0.0 | 1 | 714 | 4 | 718 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FNQ_A | 1.70e-119 | 61 | 699 | 69 | 708 | Crystalstructure of GH36 alpha-galactosidase AgaB from Geobacillus stearothermophilus [Geobacillus stearothermophilus] |
| 4FNR_A | 6.62e-119 | 69 | 699 | 74 | 708 | Crystalstructure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNR_B Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNR_C Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNR_D Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus [Geobacillus stearothermophilus] |
| 4FNP_A | 3.89e-117 | 69 | 678 | 74 | 700 | Crystalstructure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNP_B Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNP_C Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNP_D Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus [Geobacillus stearothermophilus],4FNS_A Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus],4FNS_B Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus],4FNS_C Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus],4FNS_D Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin [Geobacillus stearothermophilus] |
| 4FNT_A | 5.47e-117 | 69 | 678 | 74 | 700 | Crystalstructure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus],4FNT_B Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus],4FNT_C Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus],4FNT_D Crystal structure of GH36 alpha-galactosidase AgaA A355E D548N from Geobacillus stearothermophilus in complex with raffinose [Geobacillus stearothermophilus] |
| 4FNU_A | 5.47e-117 | 69 | 699 | 74 | 708 | Crystalstructure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus],4FNU_B Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus],4FNU_C Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus],4FNU_D Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ALJ4 | 3.63e-118 | 69 | 699 | 74 | 708 | Alpha-galactosidase AgaA OS=Geobacillus stearothermophilus OX=1422 GN=agaA PE=1 SV=1 |
| P43467 | 1.93e-109 | 62 | 716 | 72 | 732 | Alpha-galactosidase 1 OS=Pediococcus pentosaceus OX=1255 GN=agaR PE=3 SV=1 |
| Q5AU92 | 1.14e-108 | 32 | 699 | 50 | 732 | Alpha-galactosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=aglC PE=1 SV=1 |
| Q2TW69 | 1.24e-107 | 20 | 699 | 38 | 733 | Probable alpha-galactosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=aglC PE=3 SV=1 |
| B8NWY6 | 1.24e-107 | 20 | 699 | 38 | 733 | Probable alpha-galactosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=aglC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
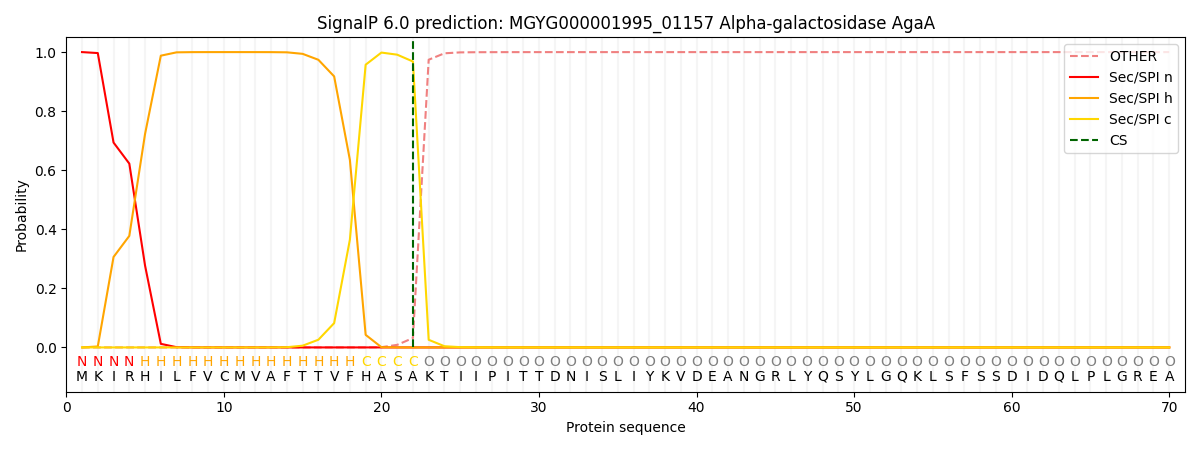
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000262 | 0.999054 | 0.000206 | 0.000166 | 0.000161 | 0.000144 |