You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002007_01808
You are here: Home > Sequence: MGYG000002007_01808
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes dispar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes dispar | |||||||||||
| CAZyme ID | MGYG000002007_01808 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 88074; End: 89066 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 197 | 324 | 1.1e-19 | 0.9032258064516129 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08522 | DUF1735 | 1.74e-19 | 36 | 149 | 3 | 114 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam00754 | F5_F8_type_C | 5.25e-13 | 201 | 317 | 8 | 117 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 2.97e-06 | 223 | 314 | 37 | 130 | Substituted updates: Jan 31, 2002 |
| pfam16391 | DUF5000 | 3.02e-06 | 215 | 290 | 8 | 90 | Domain of unknown function. This family around 200 residues locates in the C-terminal of some uncharacterized proteins in various Bacteroides and Parabacteroides species. The function of this family remains unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL06727.1 | 1.22e-250 | 1 | 330 | 1 | 330 |
| BBL03122.1 | 8.38e-233 | 1 | 330 | 1 | 330 |
| BBL15158.1 | 8.38e-233 | 1 | 330 | 1 | 330 |
| CBK65129.1 | 1.00e-157 | 1 | 330 | 1 | 330 |
| ATL46981.1 | 3.15e-64 | 16 | 330 | 21 | 329 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1W8N_A | 5.56e-07 | 152 | 328 | 430 | 596 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
| 1WCQ_A | 5.56e-07 | 152 | 328 | 430 | 596 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 2BER_A | 5.56e-07 | 152 | 328 | 430 | 596 | Y370GActive Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). [Micromonospora viridifaciens] |
| 2BZD_A | 5.56e-07 | 152 | 328 | 430 | 596 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
| 1EUT_A | 5.58e-07 | 152 | 328 | 434 | 600 | Sialidase,Large 68kd Form, Complexed With Galactose [Micromonospora viridifaciens],1EUU_A Sialidase Or Neuraminidase, Large 68kd Form [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 3.13e-06 | 152 | 328 | 476 | 642 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
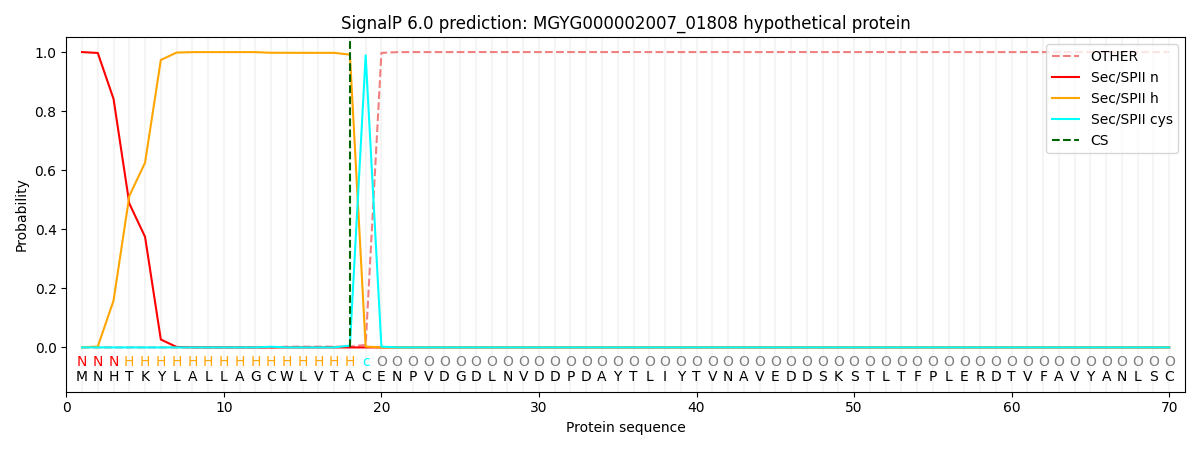
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000003 | 1.000053 | 0.000000 | 0.000000 | 0.000000 |
