You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002019_00444
You are here: Home > Sequence: MGYG000002019_00444
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
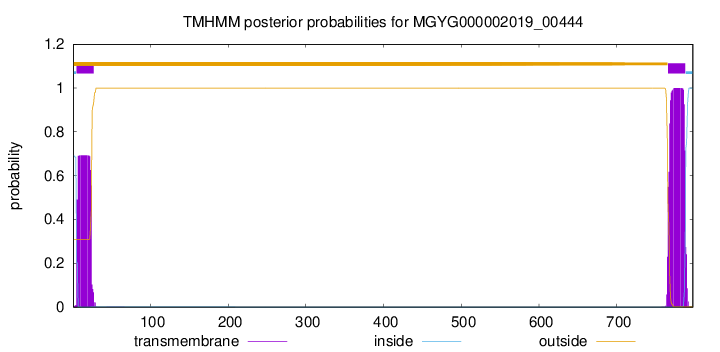
TMHMM annotations
Basic Information help
| Species | Eubacterium_R sp000431535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Eubacterium_R; Eubacterium_R sp000431535 | |||||||||||
| CAZyme ID | MGYG000002019_00444 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 113045; End: 115441 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 220 | 737 | 2.3e-94 | 0.680952380952381 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 3.96e-36 | 246 | 737 | 50 | 498 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| pfam06964 | Alpha-L-AF_C | 2.51e-34 | 546 | 731 | 1 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| smart00813 | Alpha-L-AF_C | 8.81e-25 | 546 | 731 | 1 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIL77714.1 | 3.19e-119 | 29 | 737 | 39 | 696 |
| QAA33162.1 | 1.17e-116 | 26 | 637 | 337 | 854 |
| AVR46226.1 | 5.35e-115 | 12 | 737 | 10 | 656 |
| QDA61430.1 | 5.02e-114 | 16 | 737 | 24 | 675 |
| QAA33351.1 | 1.07e-113 | 26 | 736 | 313 | 929 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 4.32e-53 | 37 | 737 | 10 | 624 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
| 6ZT6_A | 2.21e-09 | 246 | 680 | 52 | 435 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 6ZT8_A | 2.92e-09 | 246 | 680 | 52 | 435 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 2VRQ_A | 2.92e-09 | 246 | 680 | 52 | 435 | StructureOf An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_B Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_C Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus] |
| 4ATW_A | 6.52e-09 | 247 | 682 | 50 | 428 | Thecrystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_B The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_C The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_D The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_E The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_F The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 1.19e-80 | 6 | 634 | 12 | 545 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 1.22e-67 | 40 | 680 | 55 | 594 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 1.22e-63 | 29 | 682 | 42 | 593 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| Q0CTV2 | 8.60e-50 | 25 | 735 | 24 | 625 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=abfA PE=3 SV=1 |
| B8NKA3 | 1.39e-48 | 29 | 735 | 28 | 626 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abfA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
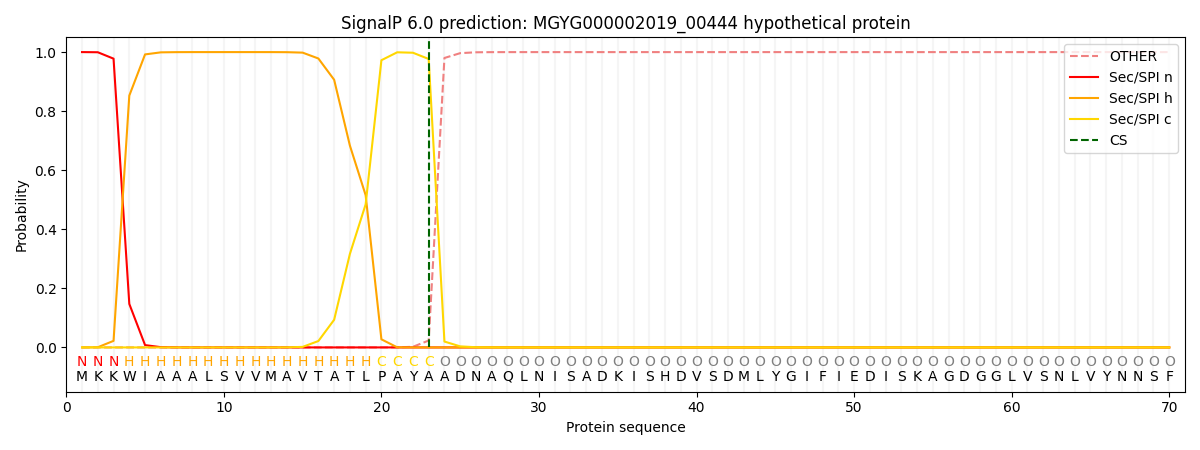
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000272 | 0.998982 | 0.000195 | 0.000188 | 0.000180 | 0.000171 |